RNA one-step-lysis and one-tube RT-PCR detection method
A technology of RT-PCR and detection method, which is applied in the field of RNA one-step cleavage and one-tube RT-PCR detection, which can solve problems such as difficult control of condition optimization, high complexity and difficulty, non-specific amplification and false positives, and achieve Improve reaction efficiency and success rate, reduce non-specific reactions, and reduce the effect of occurrence probability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
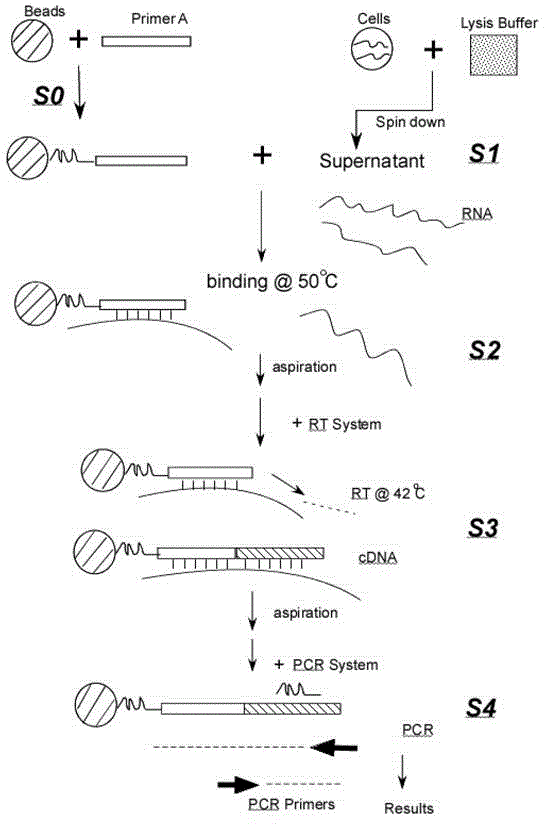
[0041] Example 1: Washing to remove genomic DNA in the supernatant of cell lysate
[0042] Primer A for detecting hTERT mRNA is Oligo(dT) 29 , PCR primer sequences are: 5'-AAGTCCTACGTCCAGTGCCAGGGGA-3' and 5'-GCTTGTTCTCCATGTCGCCGTAGC-3'.
[0043] Human lung cancer cell line A549 cells were cultured in a 24-well plate, 1000 cells / well, after overnight culture, the culture medium was aspirated, + 200ul lysate, repeatedly pipetted, transferred to a 1.5ml centrifuge tube, and shaken at room temperature for 10min , centrifuged at 4°C and 15000rpm for 20min, and the supernatant was taken to obtain the cell lysate supernatant; Mercaptoethanol, 0.1mg / ml protamine DNA (Sigma Company), the solvent is Tris-HCl with 10mmol / L and pH7.5.
[0044] Take 20ul of the supernatant of the above-mentioned cell lysate, add it to a 0.2ml PCR thin-walled tube, incubate at 50°C for 30min, then blot up the liquid in the tube, and directly add 20ul of the PCR reaction solution (containing 20ul of PCR bu...
Embodiment 2
[0049] Example 2: Immobilization of Primer A by Magnetic Beads
[0050] Primer A is Oligo(dT) 29 . PCR primers are hTERT-specific primers:
[0051] 5'-AAGTCCTACGTCCAGTGCCAGGGGA-3' and
[0052] 5'-GCTTGTTTCTCCATGTCGCCGTAGC-3'. TBST buffer 20ul, containing 10ug M-280 streptavidin magnetic beads and 2pmol biotinylated primer A (biotinylated primer A is directly modified and added to the 5' end during synthesis), put into a 0.2ml PCR thin-walled tube, room temperature for 1hr , absorb the magnetic beads in the tube wall with a magnet, blot the liquid in the tube, add 50ul TBST buffer solution, mix well, blot dry, wash repeatedly in this way for a total of 6 times, and finally blot dry; add 50ul TE buffer solution, set aside.
[0053] Cell lysis: Cell culture in 24-well plate, absorb culture medium, + 200ul lysate (same as Example 1), pipette repeatedly, transfer to 1.5ml centrifuge tube, shake at room temperature for 10min, centrifuge at 4°C and 15000rpm for 20min, take to ...
Embodiment 3
[0057] Example 3: Coupling and immobilizing primer A in direct PCR tube
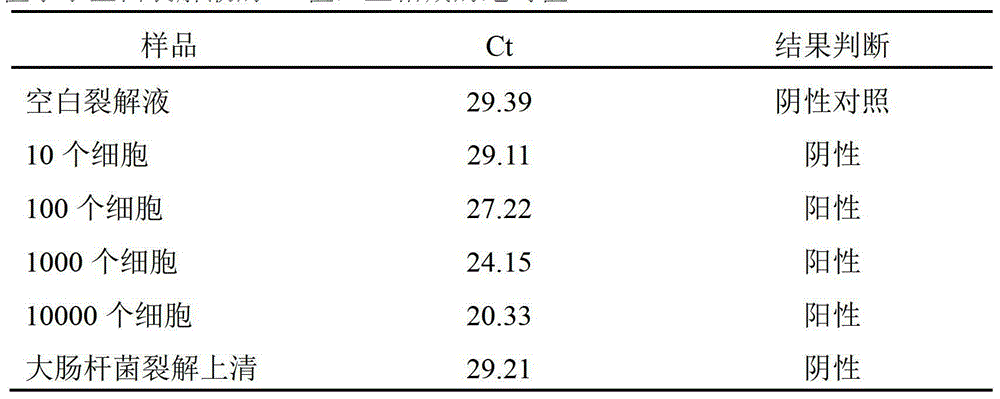
[0058] TBST buffer 50ul, containing 5pmol biotinylated Oligo(dT) 29 (See Example 2), put it into a streptavidin-coated 0.2ml PCR thin-wall tube, room temperature for 1hr, blot the liquid in the tube, add 100ul TBST buffer, mix well, blot dry, and wash repeatedly for a total of 3 Add 100ul TE buffer solution, then, blot the liquid in the tube, add 50ul cell lysate supernatant (same as Example 1), incubate at 50°C for 30min, blot the liquid in the tube, wash once with TBST buffer , add 50ul RT reaction solution (containing 50ul RT buffer, 0.5mmol / LdNTP, 10U MMLV RT enzyme), incubate at 42°C for 30min, blot the liquid in the tube, add 50ul PCR reaction solution (SYBR green I fluorescence quantification), and carry out the PCR program (Pre-denaturation at 95°C for 2min, followed by 40 cycles of 95°C for 3s and 66°C for 30s, two-step method), SYBR green I fluorescence quantitative analysis. With this method...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com