Application of Ochrobactrum sp. bacterial (H1) in quinoline degradation
A technology of quinoline and strains, applied in the direction of bacteria, microbe-based methods, microbes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] The screening of embodiment 1 bacterial strain
[0019] 1. Source and enrichment of strains
[0020] Take 5 mL of sludge from the secondary settling tank of the wastewater treatment system of Shougang Coking Plant, add 50 mL of beef extract liquid medium, and add 50 mg / L quinoline to it. Put it in a constant temperature shaker at 30°C, 150r / min, and culture for 3 days to obtain enriched culture solution.
[0021] Enrichment medium components: beef extract 3g, peptone 10g, NaCl 5g, distilled water 1000mL, pH 7.0.
[0022] Screening of strains
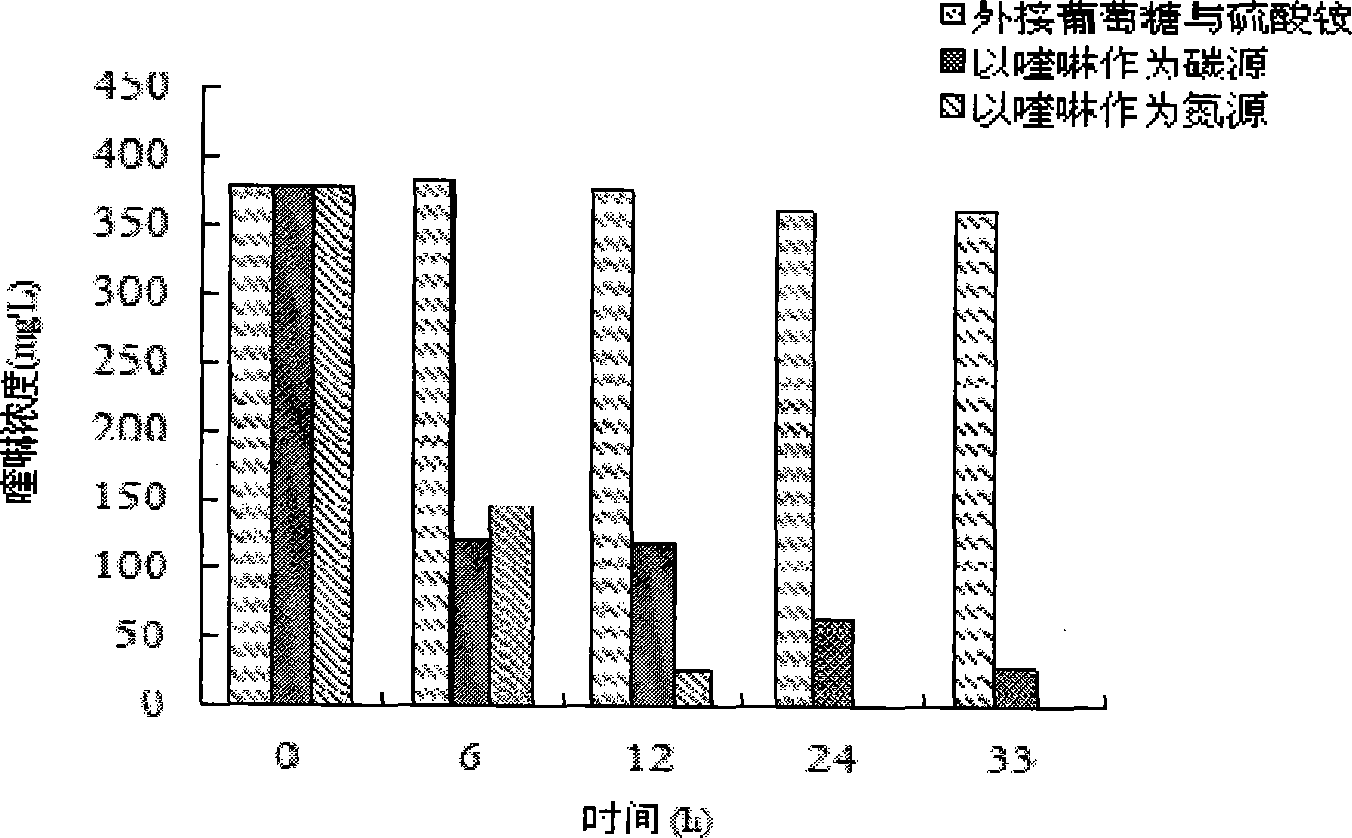
[0023] Put 5 mL of the enriched culture solution into a 10 mL centrifuge tube and centrifuge at 10000 r / min at 23 °C for 5 min in a high-speed refrigerated centrifuge. Take the precipitate and add it to normal saline to mix well, then centrifuge and repeat twice to remove residual medium and secondary metabolites. The precipitated bacterial cells obtained by centrifugation were inserted into 50 mL of the only carbon source med...
Embodiment 2
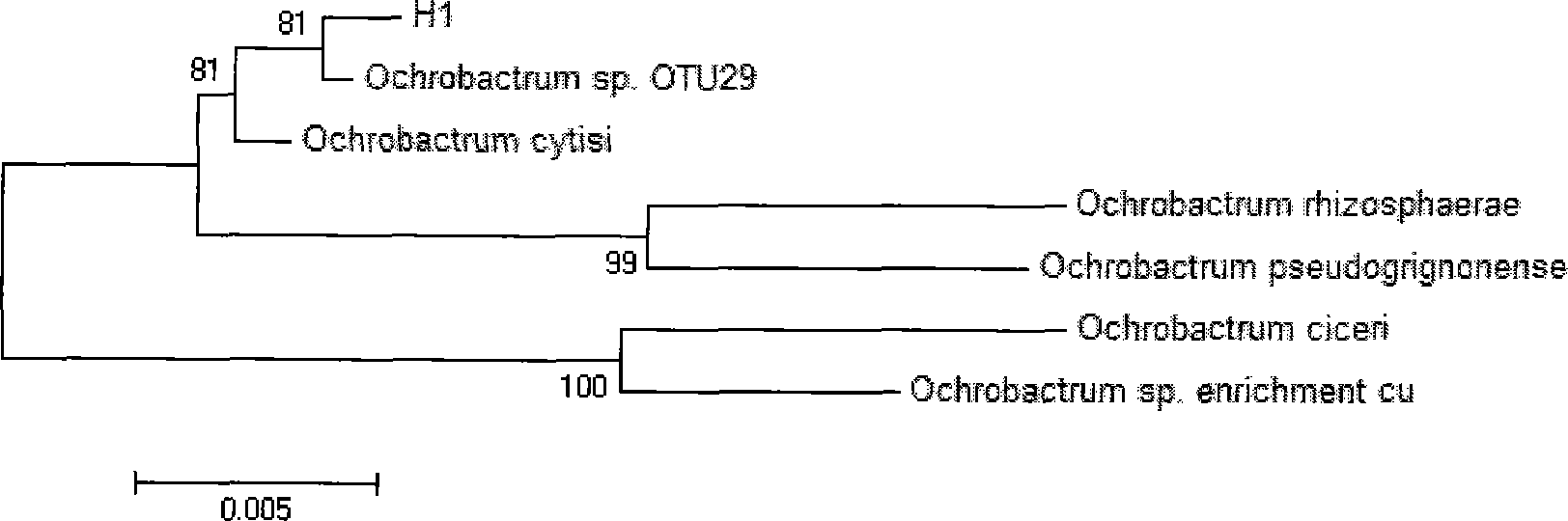
[0027] The identification of embodiment 2 bacterial strains
[0028] Using the Solarbio Bacteria Genomic DNA Extraction Kit, the genomic DNA of the strain was extracted and used as a template to perform PCR amplification using the 16SrDNA gene universal primers. Primer design for amplifying 16SrDNA:
[0029] F-primerF27: 5'-AGA GTT TGA TCC TGG CTC AG-3'
[0030] R-primer R1492: 5'-GGC TAC CTT GTT ACG ACT T-3'
[0031] Using the total DNA of the bacteria as a template, the reaction system is 50 μL:
[0032]
[0033] The PCR reaction conditions were specifically set as follows: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 1 min, annealing at 55°C for 30 s, extension at 72°C for 90 s, 40 cycles, final extension at 72°C for 5 min, and storage at 4°C. Take 2 μL of the PCR reaction product for 1% agarose gel electrophoresis experiment.
[0034] The obtained 16SrDNA sequence amplified non-purified DNA product was sequenced by Shanghai Sangon Bioengineering Tech...
Embodiment 3
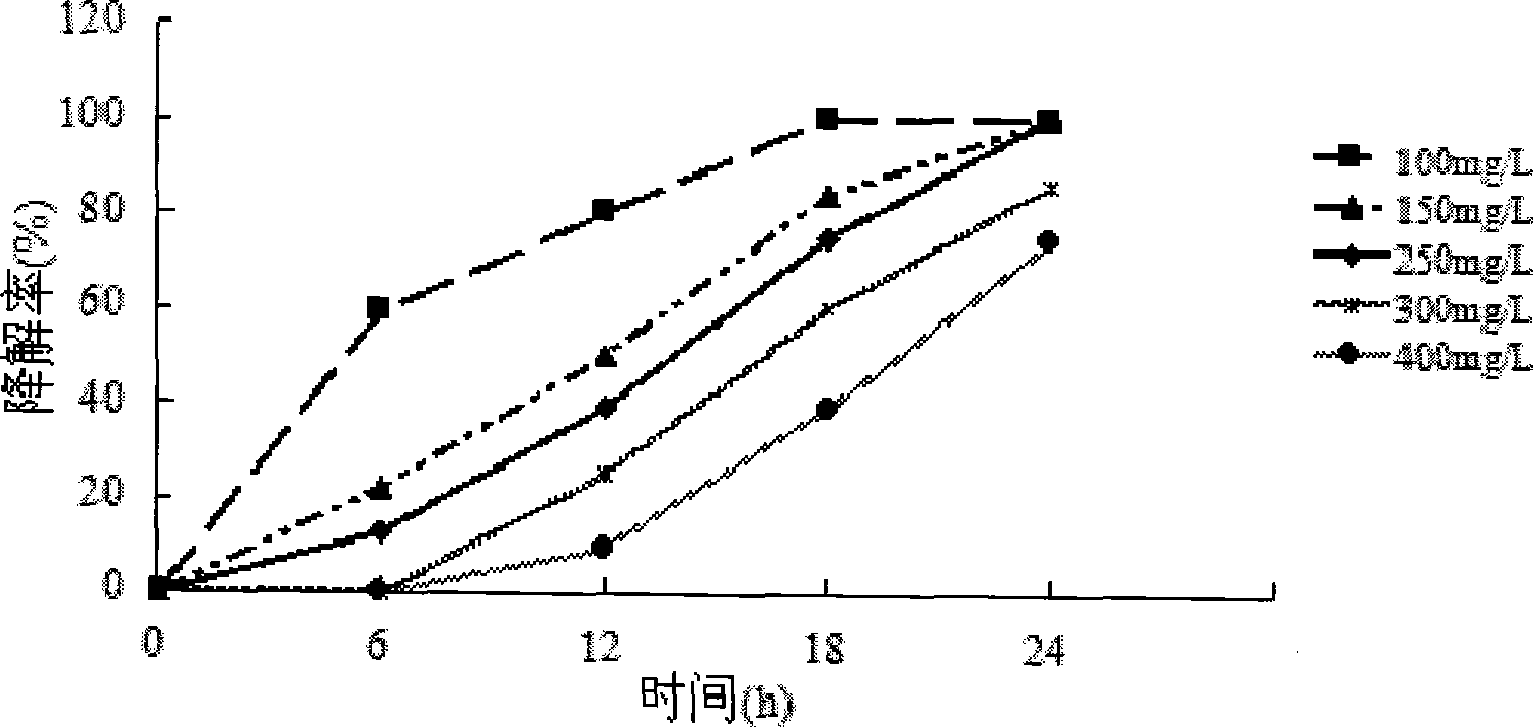
[0035] The optimal condition screening of embodiment 3 bacterial strain H1 degradation
[0036] The conditions that affect the growth of bacteria mainly include temperature, pH value, shaker speed, bacterial inoculum amount, etc. Design an L9 (3^4) orthogonal experiment according to above-mentioned four conditions, every group of experiments is set three parallel, test the degradation rate of 250mg / L quinoline in 24 hours. List the obtained test results (see Table 1). Wherein, Kn (n=1,2,3) represents the sum of the degradation rates of each factor at the same level; kn (n=1,2,3) represents the average number of corresponding K values (kn=Kn / 3); R It is the extreme difference sought for the value of k. The higher the degradation rate of the test index, the better. It can be seen directly from Table 4.1 that the test result of No. 4 test combination condition A3B1C2D3 is the largest, and it is the best effect among the nine tests. In order to find the best conditions, calc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com