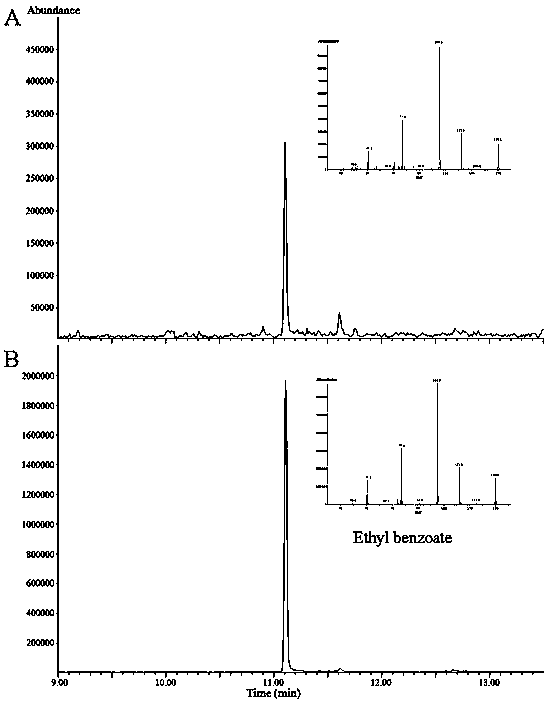
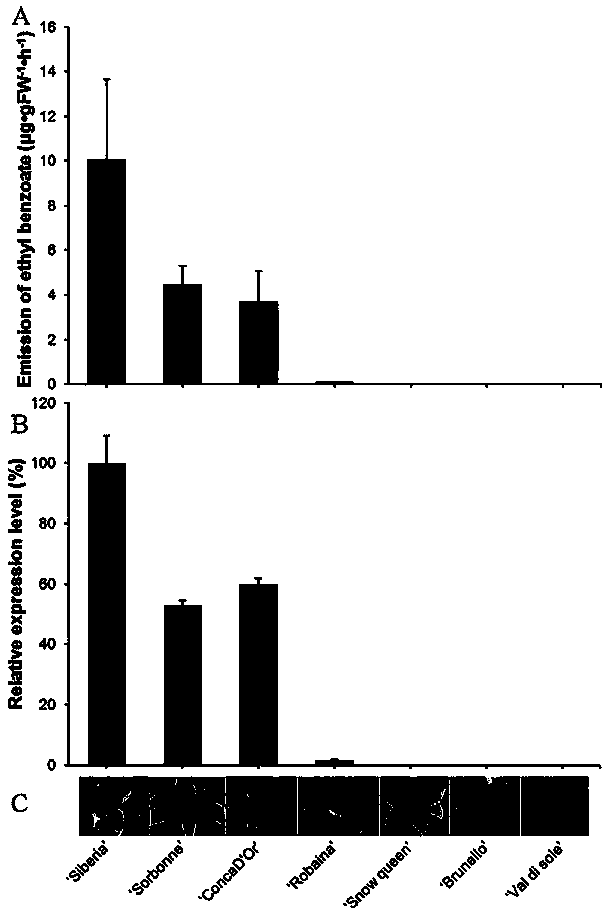
Lily ester floral gene LoAAT1 and application thereof
A lily ester and gene technology, applied in the field of genetic engineering, can solve the problem of not detecting the activity of alcohol acyltransferase and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 LoAAT1 Acquisition of gene cDNA and full-length DNA:
[0037] S1. Extraction of lily petal RNA: Weigh 0.2 g of lily petals in full bloom, add liquid nitrogen and quickly grind them into powder, and quickly transfer to 0.5 mL 2 % CTAB (containing 0.1% mercaptoethanol) extract stored at 4 °C. In a 2 mL centrifuge tube, shake until thoroughly mixed; place at room temperature for 5 minutes, lay the centrifuge tube flat to maximize the surface area; centrifuge at 12,000 rpm at 4°C for 1 min, and transfer the supernatant to a new RNase-free centrifuge tube. Add 0.1mL 5M NaCl, mix gently; add 0.3mL chloroform, mix upside down; centrifuge at 12000rpm at 4°C for 10min, transfer the upper aqueous phase to a new RNase-free centrifuge tube. Add pre-cooled isopropanol equal to the volume of the obtained water, invert and mix well, place at room temperature for 10 minutes, place the centrifuge tube horizontally to maximize the surface area; centrifuge at 12000 rpm at 4°C f...
Embodiment 2
[0045] Example 2 LoAAT1 Gene expression analysis:
[0046] Trizol method (TaKaRa) was used for RNA extraction of lily petals of different varieties, petals of different developmental stages of Siberia lily and different tissue parts of Siberia lily, and SYBR green (TaRaKa) method for fluorescence quantitative PCR. The specific principle of dye method is shown in the instruction manual. Using Primer Premier 5.0 software to design real-time fluorescent quantitative PCR primers, according to the principles of fluorescent quantitative PCR primer design, use Primer premier 5.0 to design primers respectively, and detect whether there are mismatches or primer dimers and their amplification efficiency by fluorescent quantitative PCR. Select a pair of optimal primers, P1: ATTGTGGTGCCAGTTTGCTTGC; as shown in SEQ ID NO:12. P2: CCCTTTTACCCTTCGTTGAACTTC; shown in SEQ ID NO:13. The internal reference gene ACT was designed with Primer premier 5.0 according to the design principles of R...
Embodiment 3
[0049] Example 3 Hctps1 Gene prokaryotic expression:
[0050] S1. According to the obtained LoAAT1 The full-length cDNA of the gene, containing Bam HI and not Ⅰ PCR amplification with specific primers for restriction sites. The PCR product was recovered with the Takara recovery kit, and the recovered product was directly used Bam HI and N otI restriction endonuclease for double digestion, 1% agarose gel to recover the target fragment. For pET-28a prokaryotic expression vector Bam HI and N otI restriction enzyme was used for double digestion and 1% agarose gel was used to recover large fragments. After ligation overnight at 16°C, the ligation product was transformed into Escherichia coli ( E. coli ) DH5α competent cells; the recombinant prokaryotic expression vector was obtained after the extracted plasmid was identified by enzyme digestion and sequencing.
[0051] S2. Transform Escherichia coli (Rosetta) competent cells with the identified recombinant plasmid D...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com