High-oleic-acid-content peanut molecular marker, assistant selection back cross breeding method and application of back cross breeding method
A molecular marker and backcross breeding technology, applied in the field of agricultural bioengineering, can solve the problems of high cost of fluorescent labeling, unstable results, cumbersome methods, etc., and achieve the effect of high accuracy of genotype selection, stable results, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1. High oleic acid peanut molecular marker
[0044] (1) Molecular marker primer sequence and band pattern
[0045] 1. ahFAD2B Site variant specific primers (to detect MITE insertions)
[0046] Design the forward primer MITE-INS-F at 63 bp of the MITE insertion sequence of the peanut FAD2 mutant gene, and design the reverse primer MITE-INS-R at the downstream 472 bp of the MITE insertion to detect the MITE insertion; if there is a site mutation, the expected band of 613 bp will be amplified; if there is no site variation, no band will be amplified, and the primers obtained are as follows:
[0047] Forward primer MITE-INS-F: 5’……GGATGATGGATTGTATGG…….3’
[0048] Reverse primer MITE-INS-R: 5'.....CTCTGACTATGCATCAG...3'.
[0049] 2. ahFAD2B Site-free specific primers (detection of wild-type without MITE insertion)
[0050] Design the forward primer WILD-F at -80 bp of the peanut FAD2 wild-type gene, and design the reverse primer WILD-R at 654 bp. If there ...
Embodiment 2
[0058] Example 2. Genotyping and Verification of Oleic Acid Content Using Molecular Markers
[0059] 1. Genotyping and oleic acid content of Yuanza 9102×wt09-0023 offspring
[0060] The DNA of the F3:4 individual offspring of the Yuanza 9102×wt09-0023 hybrid combination was extracted, and the above-mentioned designed ahFAD2B The two pairs of molecular marker primers at the locus were amplified by PCR respectively, and the band patterns of the amplification results were as follows: figure 1 , figure 2 (The three-digit strain number in the figure is Yuanza 9102×wt09-0023, and the four-digit strain number is Fuhua 12×wt09-0023). Combined with the sequencing results of FAD2A, the band type of FAD2B was compared with the genotype and oleic acid content (Table 2).
[0061] The results showed that under the AA background, the genotypes of the B genome changed from BB, Bb to bb, and the oleic acid content increased from 31.9%, 38.1% to 63.3%; The acid content increased from 3...
Embodiment 3
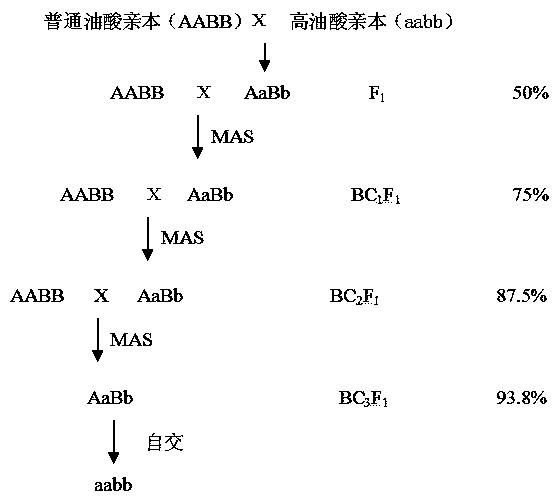
[0069] Example 3: Backcross Breeding Method of High Oleic Acid Peanut Using Molecular Markers , see image 3 , the method includes the following steps:
[0070] (1) Use parents with ordinary oleic acid content (oleic acid content 70%) were crossed to get F 1 ;
[0071] (2) Select offspring F 1 As the male parent, backcross with the peanut parent of genotype AABB to obtain the offspring BC 1 f 1 ;
[0072] (3) Extract peanut BC 1 f 1 For the DNA of a single plant, first use a pair of primers in FAD2A to amplify the FAD2A gene fragment, wherein the forward primer is aF19: GATTACTGATTATTGACTT, and the reverse primer is R3: CCCTGGTGGATTGTTCA,
[0073] Sequencing after amplification to select a single plant containing the Aa genotype;
[0074] Then use the peanut ahFAD2B The mutation-specific primers and non-variation-specific primers at the locus were used to perform PCR detection on the individual plants containing the Aa genotype, and the two markers were dominant,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com