Kit used for rapid detection of enterobacter sakazakii in milk, and applications thereof
A technology of Enterobacter sakazakii and a kit, which is applied in the direction of microorganism-based methods, measurement/inspection of microorganisms, microorganisms, etc., can solve the problems of unsuitable on-site detection, errors in naked eye observation, time-consuming and labor-intensive, etc., and achieve suitable on-site detection, The effect of short time consumption and simple equipment requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] 1. DNA extraction of tested milk samples: Add 1 mL of DNA extraction solution (0.1 mol / L Tris-HCl, 0.1 mol / L ethylenediaminetetraacetic acid EDTA, 0.1 mol / L to 1 mL of tested milk. L Sodium Phosphate Na 3 PO 4 , 1.5mol / L sodium chloride NaCl, 1% CTAB hexadecyltrimethylammonium bromide, pH8.0), 10 μL proteinase K, 10 μL lysozyme, 10 μL lysozyme at 37°C at 200r / min Incubate at rotational speed for 1.5 hours; add 200 μL of 20% SDS to it and incubate at 65°C for 1 hour; add an equal volume of chloroform and centrifuge at 13,000 r / min for 1 minute and take the supernatant; add 0.6 times the volume to the obtained supernatant to pre-cool Mix the isopropanol upside down; transfer it to the adsorption column AC and centrifuge at 10,000r / min for 30 seconds, then add 700μL of rinsing solution; centrifuge at 10,000r / min for 2 minutes, pour off the liquid, add 500μL of rinsing solution, and centrifuge to remove the rinsing solution as much as possible ; Add 50 μL of elution buffe...
Embodiment 2
[0031] 1. The DNA extraction method of the tested milk sample is the same as in Example 1
[0032] 2. Use the extract as a template for LAMP amplification, and the reaction system is a 50 μL reaction system:
[0033]
[0034]
[0035] LAMP amplification conditions are: react at 95°C for 5 minutes, quickly place at 2°C and add Bst enzyme, react at 63°C for 60 minutes, and passivate at 80°C for 5 minutes.
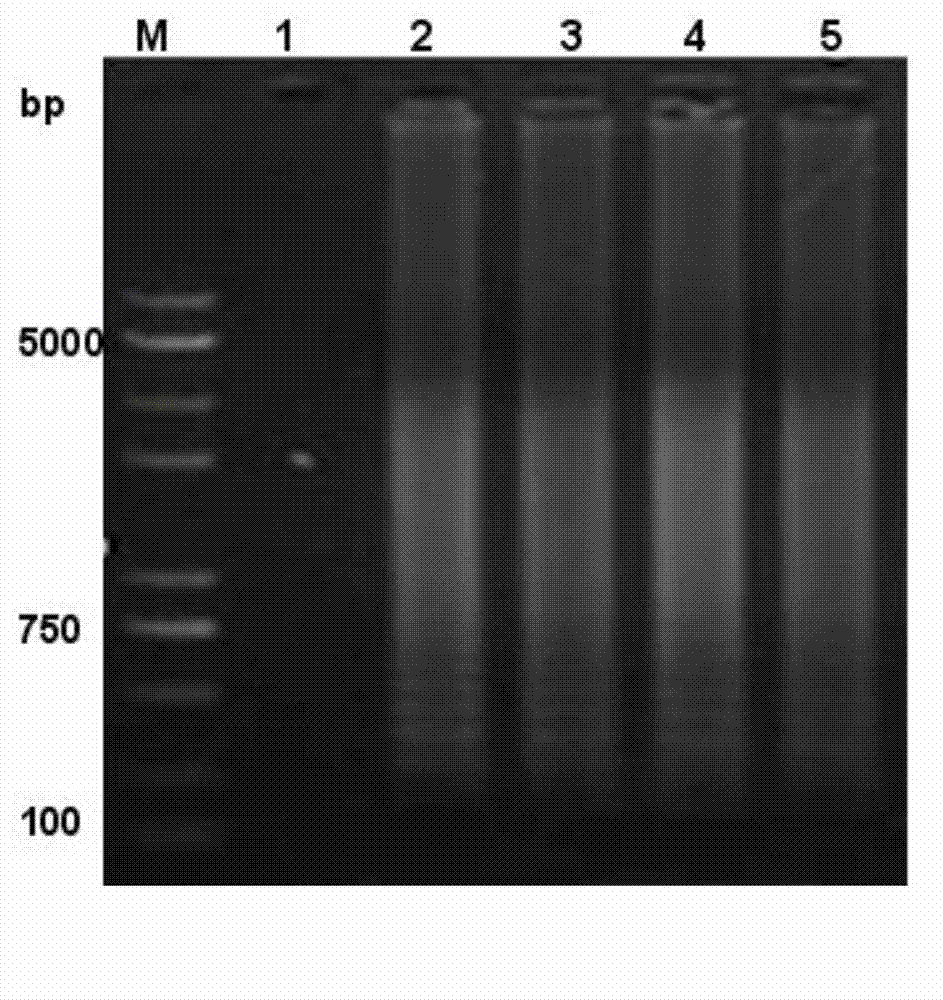
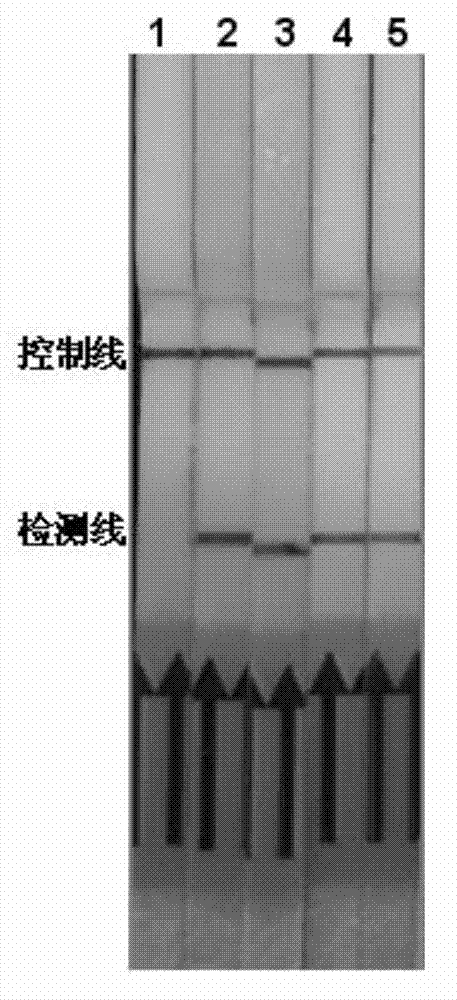
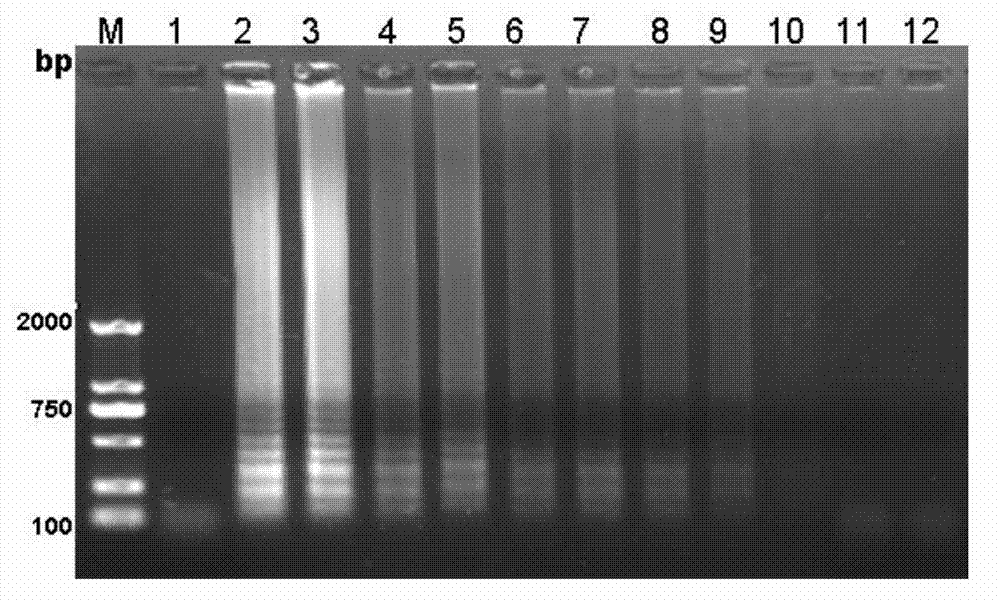
[0036] 3. Test strip detection of amplified products: Take 2 μL of probe 5'-CGAAGACTCTCGCACTCGCAGCA-3' and hybridize with the product at 64°C for 5 minutes, then take 10 μL of the hybridization product and incubate with 110 μL buffer for 5 minutes, and finally put the test strip Put it in and incubate for 15 minutes, observe whether there is a band on the detection line of the test strip for result interpretation. The result is as figure 1 , 2 As shown, the blank is not amplified and there is no band in the detection line.
[0037] This implementation method has simpl...
example 3
[0038] Detection of Enterobacter sakazakii in example 3 milk
[0039] 1. The DNA extraction method of the tested milk sample is the same as in Example 1
[0040] 2. Use the extract as a template for LAMP amplification, and the reaction system is a 50 μL reaction system:
[0041]
[0042]
[0043] LAMP amplification conditions are: react at 90°C for 10 minutes, quickly place at 4°C and add Bst enzyme, react at 65°C for 40 minutes, and passivate at 90°C for 5 minutes.
[0044] 3. Test strip detection of the amplified product: Take 2 μL of probe 5’-CTAGGTCACACGAAGACTGCAGC-3’ and hybridize with the product at 64°C for 5 minutes, then take 10 μL of the hybridization product and incubate with 110 μL buffer for 5 minutes, and finally put the test strip Put it in and incubate for 15 minutes, observe whether there is a band on the detection line of the test strip for result interpretation. The result is as figure 1 , 2 As shown, the blank is not amplified and there is no band...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com