Recombination gene and method for increasing aspergillus niger expressed saccharifying enzyme
A recombinant gene, Aspergillus niger technology, applied in the field of genetic engineering, can solve the problems of unpredictable results, a lot of time and labor costs, time-consuming and laborious Aspergillus screening, etc., and achieves transformation efficiency and expression of saccharification enzyme activity. Eliminate the need for screening and transformation effect of subprocess
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
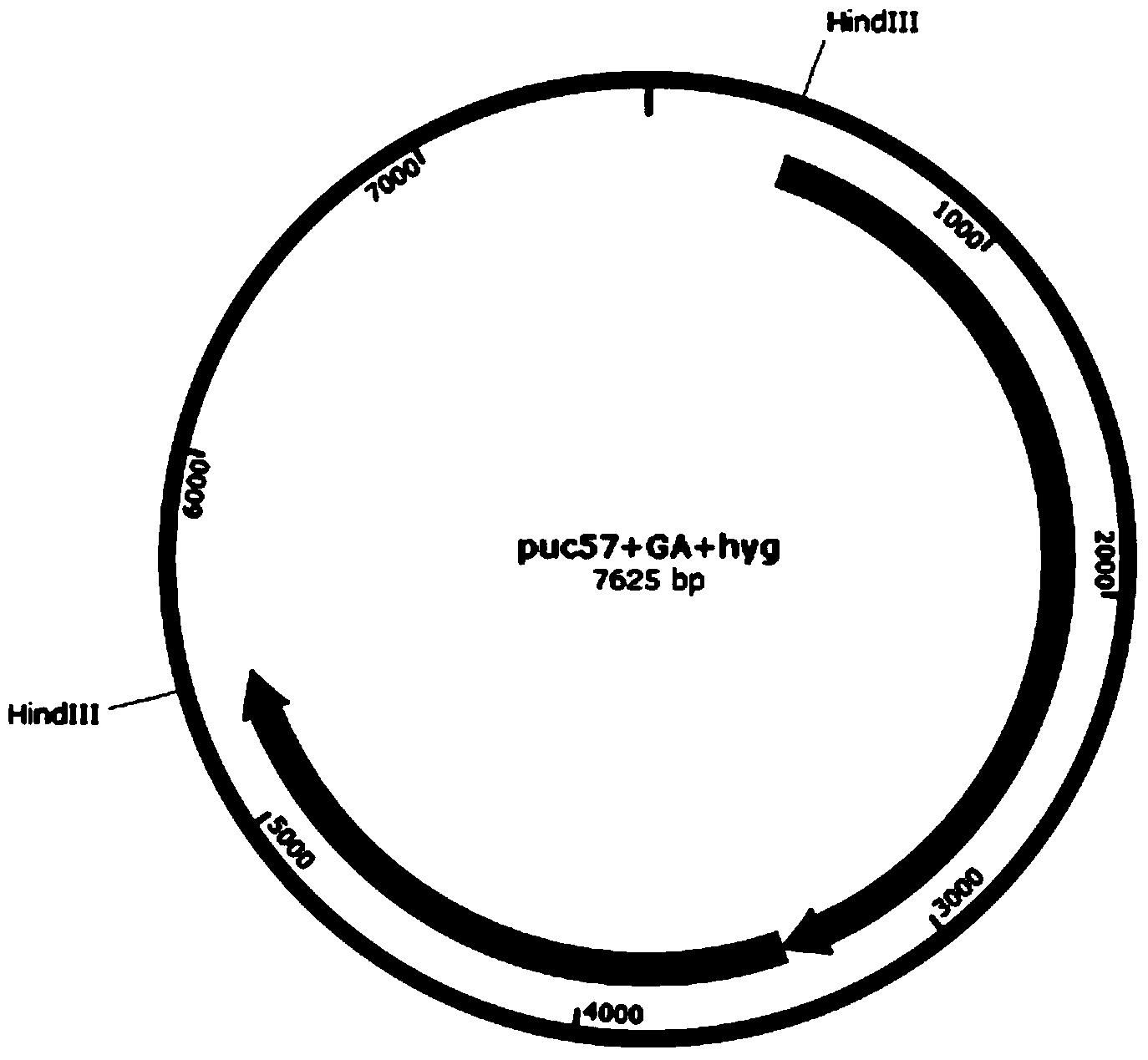
[0037] Example 1 Randomly integrated GA expression cassette construction
[0038] In order to compare with the site-specific integration results, a random integration plasmid was constructed, which contains the following parts:
[0039] (1) Fragments obtained after EcoRI-HindIII double digestion of pUC57 plasmid;
[0040] (2) Glucoamylase (TeGA) expression cassette, wherein the glucoamylase is a Talaromyces variant, synthesized by GenScript, and its sequence is shown in SEQ ID NO.1;
[0041] (3) Selection marker Hyg expression cassette, synthesized by GenScript Company, the sequence is shown in SEQ ID NO.2;
[0042] First, use primers GA_F and GA_R, hyg_F0 and hyg_R0 to amplify the GA gene and hyg gene with recombination arms by PCR, and then use PCR to assemble the GA gene and hyg gene into a straight chain through the overlapping sequences at both ends of the primers GA_F and hyg_R0 The fragment was then recombined with the pUC57EcoRI-HindIII digested linear recovery fragm...
Embodiment 2
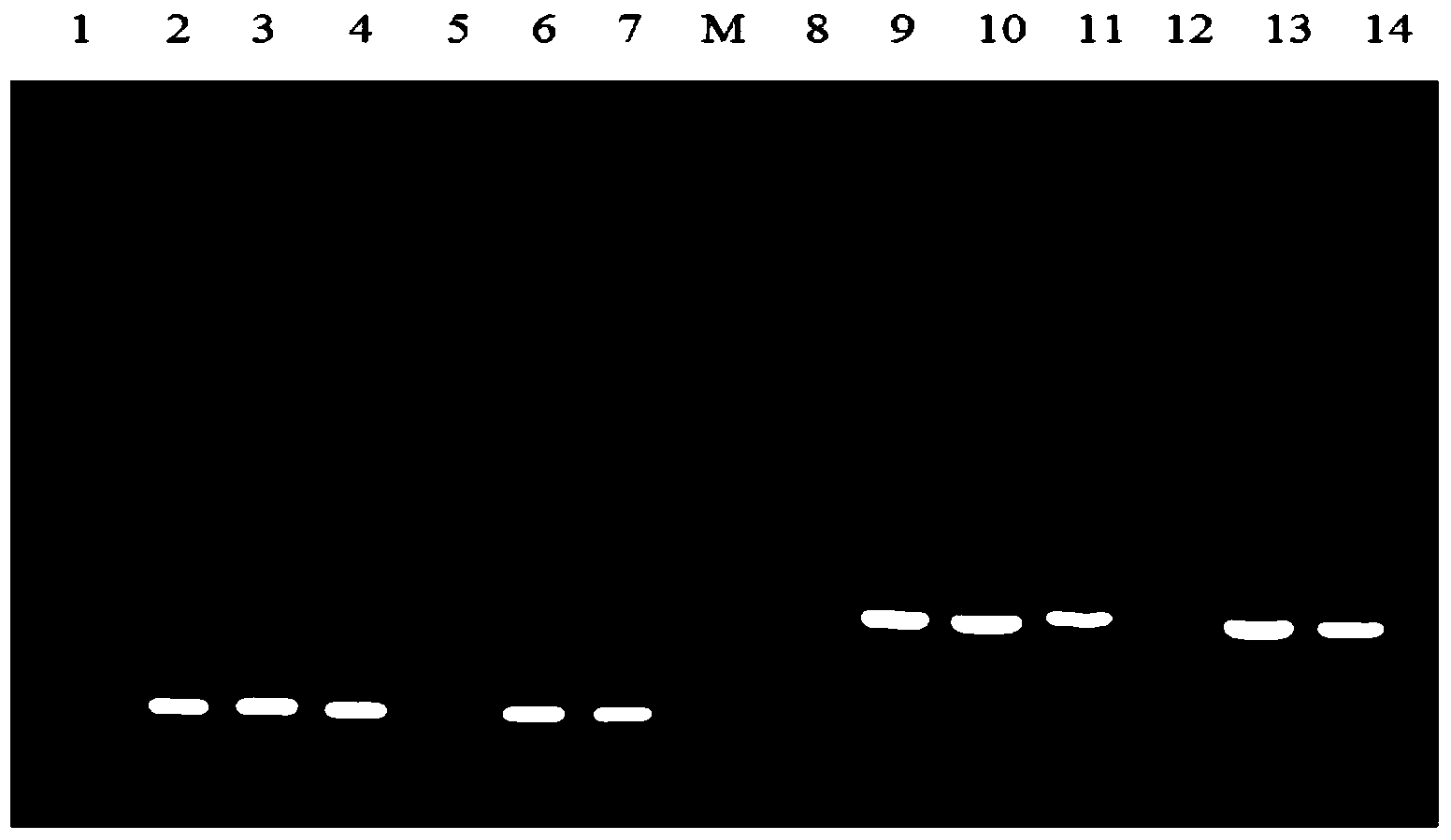
[0045] Embodiment 2 Transformation and screening of randomly integrated GA
[0046] The preparation of protoplast: cultivate Aspergillus niger mycelia (Aspergillus niger CBS513.88, purchased from the Netherlands Fungal Culture Collection (CBS-KNAW Fungal Biodiversity Centre). The mycelium was filtered from the culture medium through mira-cloth (Calbiochem Company) and washed with 0.7M NaCl (pH 5.8). Sigma) and lysozyme (Sigma) 0.2% enzymatic solution. 30°C, 65rpm enzymatic hydrolysis for 2.5-3h. The enzymatic hydrolysis containing protoplasts was then placed on ice and filtered through four layers of lens tissue. The obtained filtrate was centrifuged at 3000rpm at 4°C for 10min, and the supernatant was discarded; the protoplasts attached to the tube wall were treated with STC solution (1M D-sorbitol, 50mM CaCl 2 , 10mM Tris-HCl, pH7.5) washed once, and finally the protoplasts were resuspended in an appropriate amount of STC solution.
[0047] Protoplast transformation: Ad...
Embodiment 3
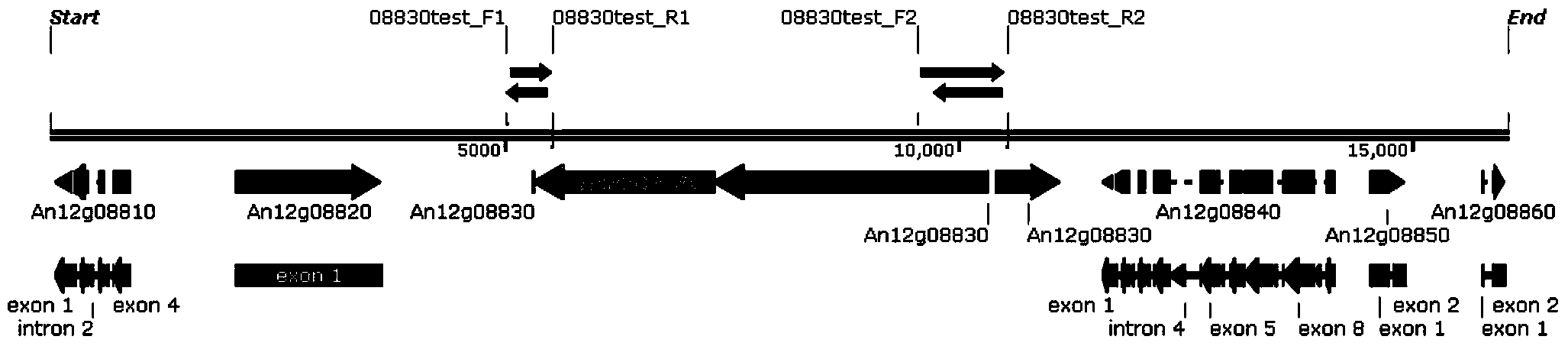
[0048] Example 3 Construction of site-specific integration of GA expression cassettes
[0049] Site-specific integration was performed according to the method described by Kim (Kim et al., Biochem. Biophys. Res. Commun390(3):983-988, 2009) et al. (Double-Joint PCR with Split Dominant Selectable Markers). Two exogenous DNA fragments are required for co-transformation into Aspergillus niger:
[0050] Fragment 1: An12g08830 locus 5' flanking sequence (SEQ ID NO.3)+hyg2 (SEQ ID NO.4)
[0051] Fragment 2: An12g08830 locus 3' flanking sequence (SEQ ID NO.5)+TeGA expression cassette (SEQ ID NO.1)+hyg1 (SEQ ID NO.6)
[0052] Among them, hyg1 contains a promoter and a 680bp part of the hyg CDS sequence, and hyg2 contains another part of the hyg CDS sequence (616bp of which is homologous to the hyg part of the CDS sequence in hyg1) and a terminator.
[0053] Construction of Fragment 1: Genomic DNA of Aspergillus niger CBS513.88 (purchased from CBS-KNAW Fungal Biodiversity Centre) was ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com