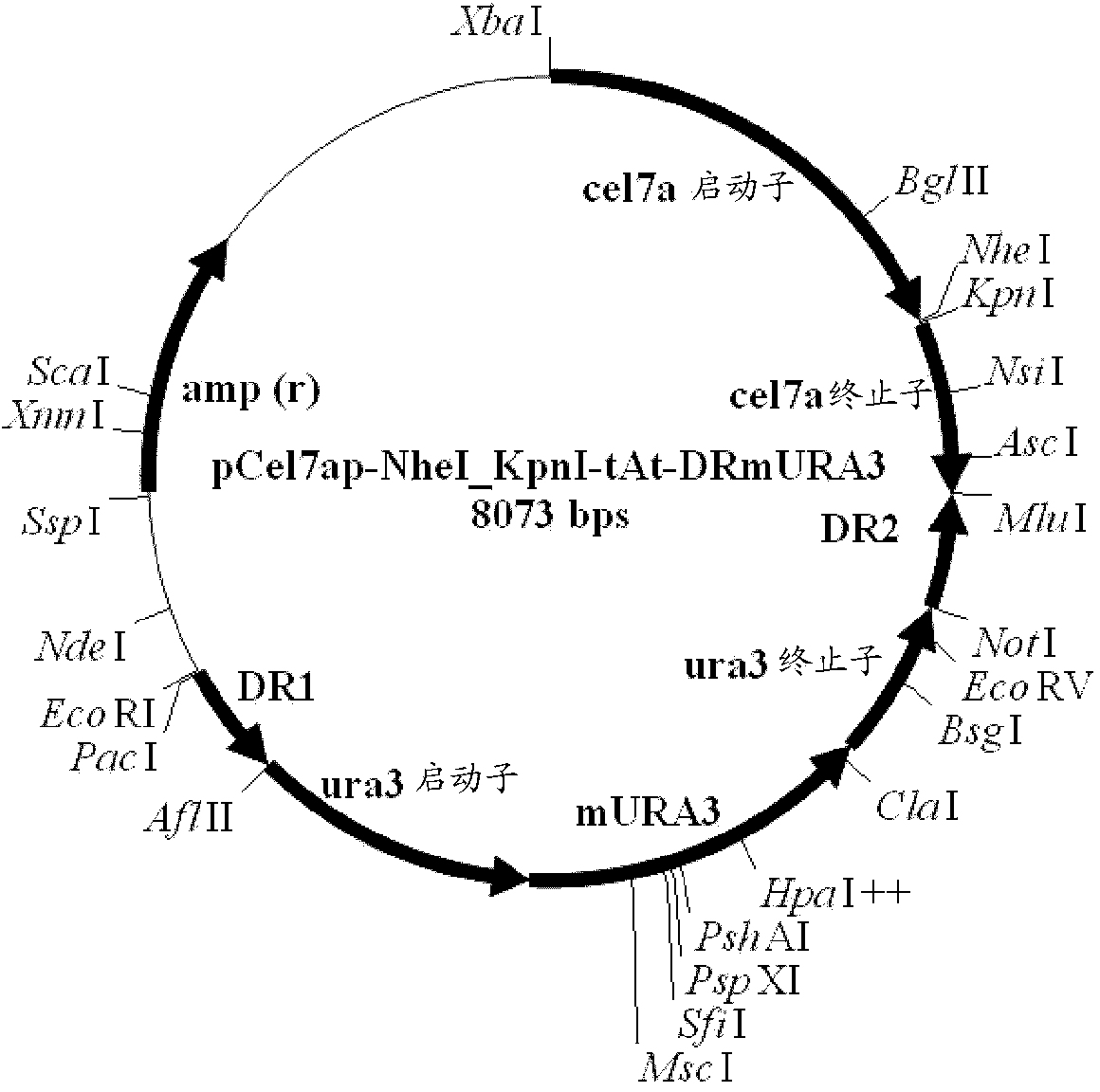
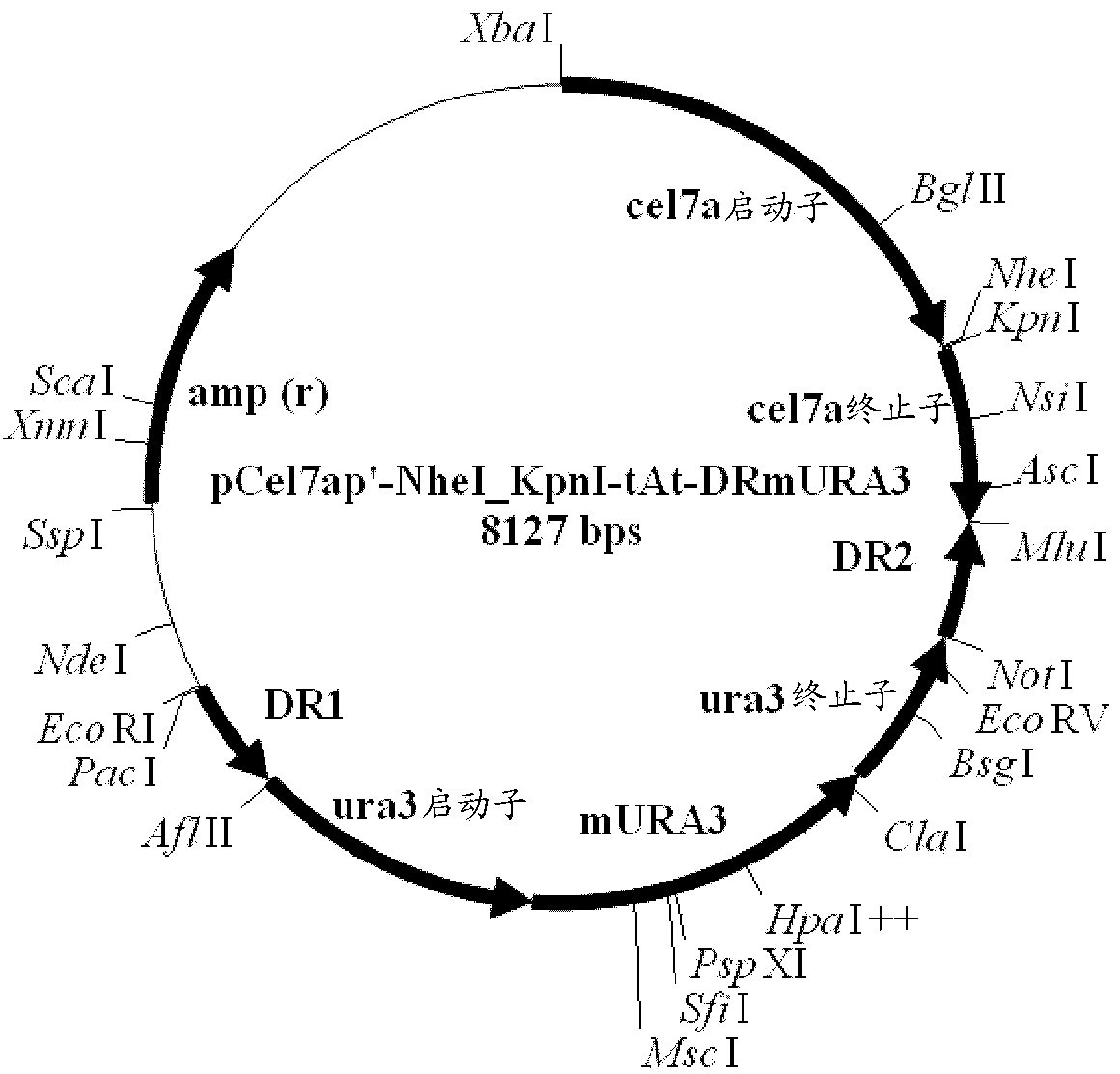
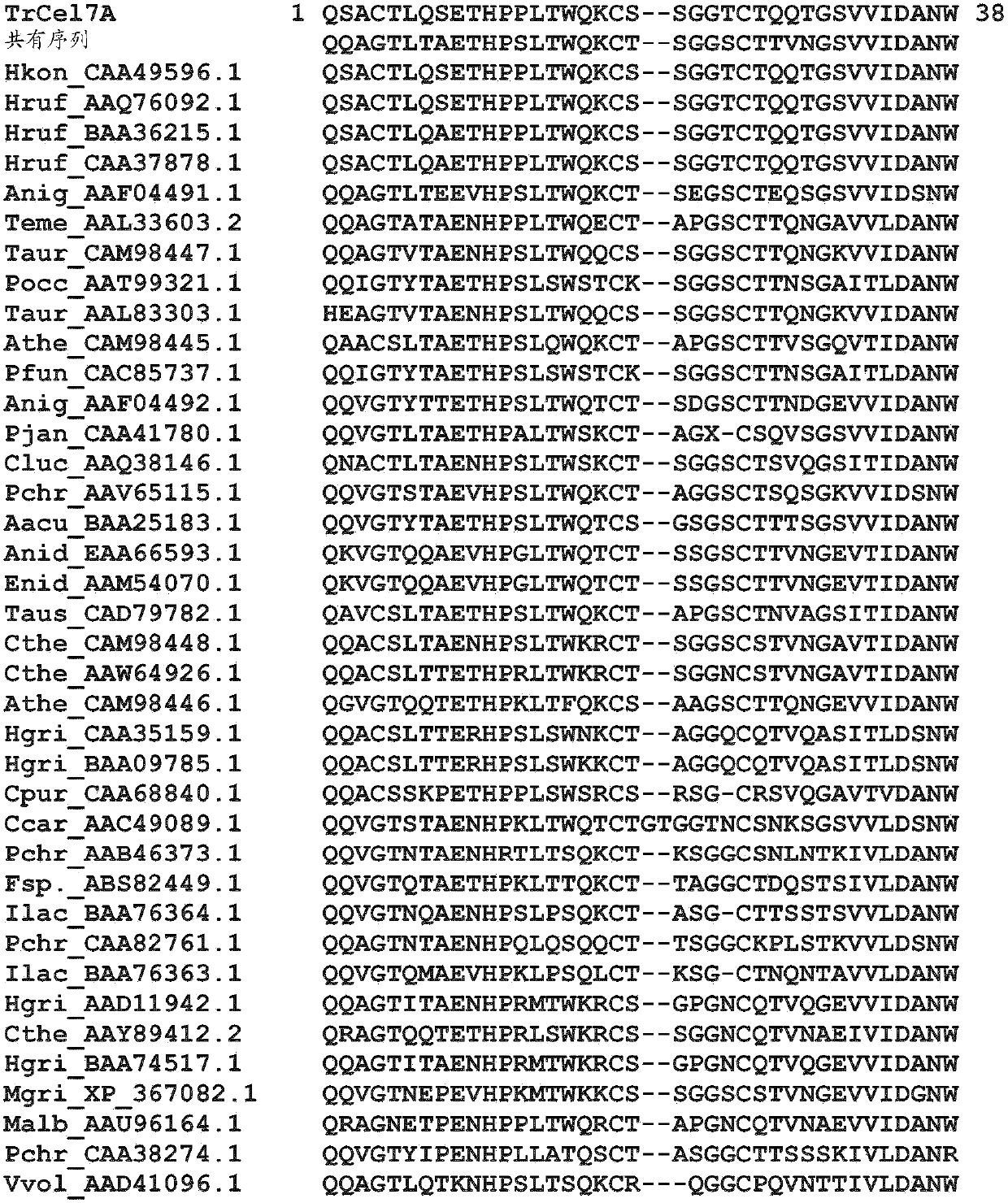
Novel cellobiohydrolase enzymes
A cellobiose, hydrolase technology, applied in hydrolase, glycosylase, enzyme and other directions, can solve the problem of irrelevance of performance and activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0188] Example 1 describes the strains and vectors used in the following examples. Example 2 describes the cloning of the TrCel7A gene and the preparation of a site-saturation mutagenesis library of TrCel7A. Example 3 describes the preparation of a random mutagenesis library of TrCel7A. Example 4 describes the transformation of a T. reesei host strain with a genetic construct expressing an isolated cellobiohydrolase. Example 5 describes the expression of wild-type and isolated TrCel7A cellobiohydrolase from microcultures. Example 6 describes the sequencing of polynucleotides encoding isolated cellobiohydrolases. Examples 7 to 12 describe high-throughput screening assays for identifying isolated cellobiohydrolases with improved activity on processing substrates.
[0189] Example 1: Strains and vectors
[0190] E. coli strain DH5α (F-ф80lacZΔM15Δ(lacZYA-argF)U169recA1endA1hsdR17(rk-,mk+)phoA supE44thi-1gyrA96relA1λ-) was obtained from Invitrogen (cat. No. 18265-017 and / or 18...
Embodiment 2
[0192] Example 2: Site Saturation Mutagenesis (SSM) of TrCel7A
[0193] a. SSM PCR
[0194] A site-saturation mutagenesis (SSM) library of TrCel7A was generated using primers containing degenerate codons (NNS) targeting multiple amino acid positions. SSM was performed using a two-step PCR method involving Megaprimer synthesis followed by PCR-mediated overlap extension. Use iProof TM High-fidelity DNA polymerase (Bio-Rad) was used for PCR reactions. A vector containing genomic DNA encoding TrCel7A with flanking sequences (SEQ ID NO: 384) was used as a template for the L375X library. A vector containing genomic DNA encoding TrCel7A-R449E-R450E with flanking sequences (SEQ ID NO:385) was used as template for all other SSM libraries.
[0195] For each SSM library, MegaPrimer A was amplified using the outer forward primer AC639 and the inner mutagenic reverse primer, while MegaPrimer B was generated by combining the outer reverse primer AC640 with the inner mutagenic forward pr...
Embodiment 3
[0205] Example 3: Random mutagenesis of TrCel7A
[0206] a. Error-prone PCR
[0207] Use by error-prone PCR II DNA polymerase (Agilent) generated random mutagenesis libraries. Two error-prone PCRs were performed. One reaction used 20 fmol of the vector containing the polynucleotide encoding TrCel7A-R449E-R450E and flanking sequences (SEQ ID NO: 442). Another error-prone PCR was performed using 100 fmol of the vector containing the polynucleotide encoding TrCel7A and flanking sequences (SEQ ID NO: 443). Both reactions contain II DNA polymerase and primers AC639 and AC640. The annealing temperature was set at 60 °C and continued for 20 cycles to complete the amplification. Use Wizard Error-prone PCR amplicons were subjected to agarose gel electrophoresis using the Gel and PCR Purification System (Promega), and approximately 1.6 kb amplicons were purified from the gel. The primer sequences are shown below:
[0208] AC639: 5'CCGCACTCCCCATCCCCTACCATCTATCCGAACTTCCCCGTC (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com