Construction method and application of BL21(DE3)delta aroA strain
A construction method and technology of BL21, applied in the field of construction of BL21ΔaroA strain, can solve the problem of difficult target gene homologous replacement and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
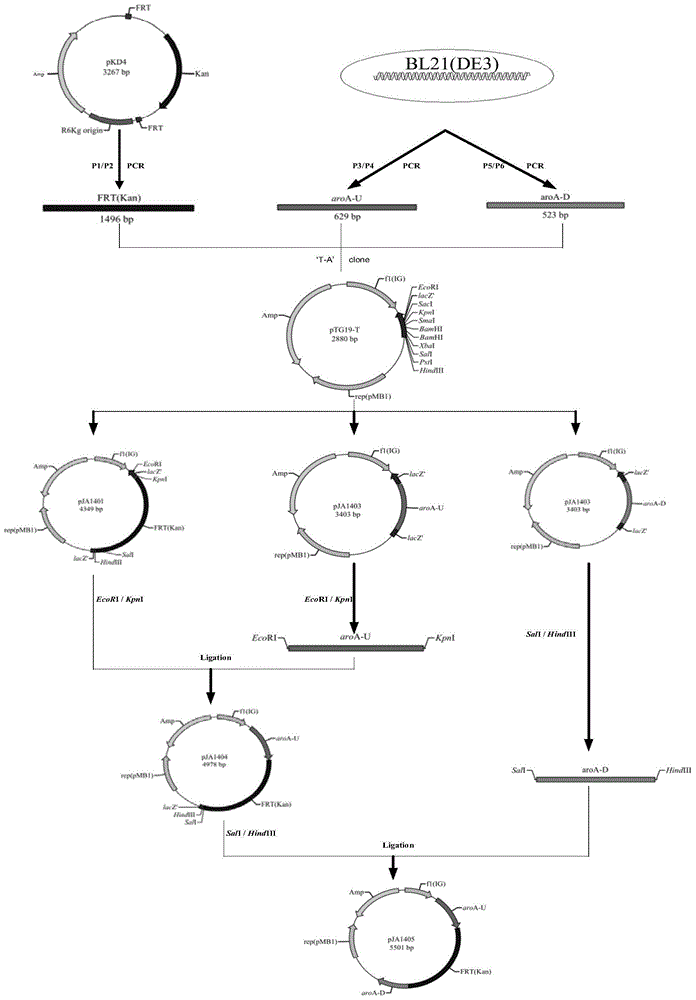
[0066] Such as figure 1 As shown, construct the aroA gene knockout targeting fragment
[0067] (1) Construction of pJA1401 plasmid
[0068] Using the pKD4 plasmid as a template, use primers P1 and P2 for PCR amplification. The PCR reaction system is (25 μl): 10×EasyPfu Buffer 2.5 μl, 2.5mM dNTPs 2.5 μl, P1 (20 μM) 0.5 μl, P2 (20 μM) 0.5 μl , pKD4 (2.5ng / μl) 1μl, EasyPfu DNA Polymerase 0.5μl, ddH 2 O 17.5 μl;
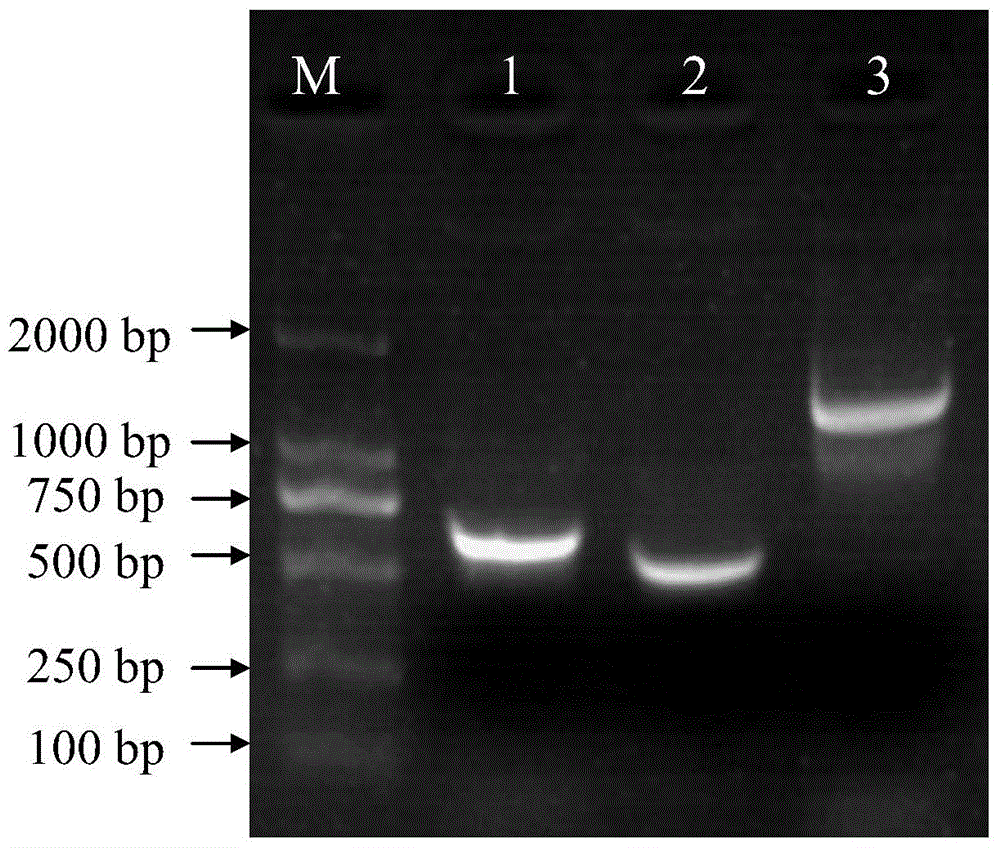
[0069] The reaction parameters were as follows: after pre-denaturation at 94°C for 3 min, PCR amplification was performed according to the following procedure: denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 2 min and 30 s, a total of 33 cycles, and finally extension at 72°C for 5 min. After the reaction was completed, agarose gel electrophoresis was carried out to obtain figure 2 , figure 2 Middle, M: D2000 Marker; Lane 1: aroA-U(aroA E.coli upstream sequence) fragment; Lane 2: aroA-D (aroA E.coli downstream sequence) fragment; ...
Embodiment 2
[0092] Obtaining BL21(DE3)ΔaroA Strain
[0093] The plasmid pKD46 was transformed into BL21(DE3) competent cells by heat shock method, and positive monoclonal colonies were screened on LB medium containing Ap resistance at 30°C. Pick a single colony and incubate in 1ml (1.5ml eppendorf centrifuge tube) LB liquid medium containing Ap with shaking at 30°C for 7h, then add all 1ml of bacterial liquid into 50ml (250ml Erlenmeyer flask) LB liquid medium containing Ap and shake at 30°C Incubate for 4 hours, add arabinose at a final concentration of 0.2% and culture for another 1.5 hours, and then collect the bacteria to prepare electric shock competent cells BL21(DE3) / pKD46.
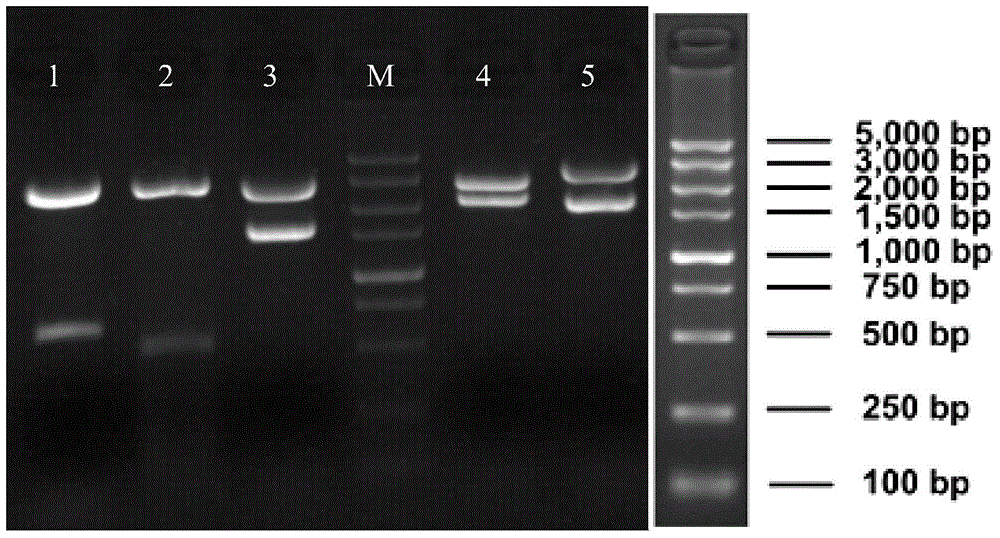
[0094] The aroA gene knockout target fragment recovered in Example 1 was transformed into competent cells of the BL21(DE3) / pKD46 strain by electric shock, firstly incubated with shaking at 30°C for 1 hour, and then incubated with shaking at 37°C for 1.5 hours, then collected the bacteria and spread evenly On ...
Embodiment 3
[0098] BLA - Obtaining of (BL21(DE3)ΔaroA) strain
[0099] Elimination of Kan resistance gene in BL21(DE3)ΔaroA strain
[0100] Prepare BL21(DE3)ΔaroA strain electroshock competent cells, transform the plasmid pCP20 into BL21(DE3)ΔaroA competent cells by electric shock, resuscitate the cells with 1ml of LB liquid medium added with Ap, incubate at 28°C for 4h, and collect the cells , and then uniformly spread on LB solid medium containing Ap, cultured overnight at 28°C to screen positive single colonies.
[0101] Pick a single colony and add it to 700μl (1.5ml eppendorf centrifuge tube) liquid LB medium, shake and incubate at 42°C for 2.5h, then take 100μl of the bacterial liquid and spread it evenly on the surface of the LB solid medium, cultivate overnight at 42°C, and screen the single colony. When the single colony is the BLA that the Kan resistance gene has been eliminated - (BL21(DE3)ΔaroA) strain.
[0102] BLA obtained for verification - (BL21(DE3)ΔaroA) strain Kan ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com