A rapid and sensitive method for the detection of small RNAs
A sensitive detection, small molecule technology, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of not showing bands, background interference, long time, etc., to shorten the detection time and improve the detection sensitivity , The effect of shortening the detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] In this example, the detection of Osa-miR156 in rice leaves, let-7-a in mouse hepatocytes, and chicken blood was taken as an example.
[0038] The flow chart of the whole detection method is as follows: figure 1 shown.
[0039] In the process of realizing the object of the present invention, the following steps are usually included:
[0040] 1. Material selection: fresh animal and plant tissues, blood, or fresh -70°C frozen samples.
[0041] 2. Reagent configuration:
[0042] 1) DEPC treated water RNase free water: deionized water is treated by Millpore water processor, add 1 / 1000 DEPC water after treatment, sterilize under high temperature and high pressure for 30 minutes, and cool to room temperature;
[0043] 2) TBE: Tris: 108g; Na 2 EDTA·2H 2 O: 7.44g; boric acid: 55g, add 800ml of deionized water, stir to dissolve, add deionized water to 1L, store at room temperature;
[0044] 3) 10% APS: 1g ammonium persulfate, dissolved in 10ml double distilled water;
[...
Embodiment 2
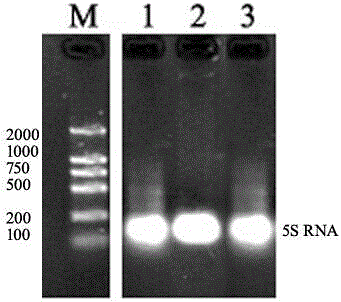
[0083] Detection sensitivity test
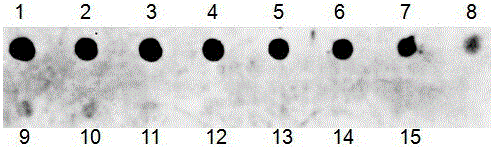
[0084] This embodiment is based on the method of Example 1. During liquid phase hybridization, Osa-miR156 in rice leaves with different concentrations was used for liquid phase hybridization and solid phase detection, and the detection sensitivity test was carried out. The results are as follows: Figure 4 as shown, Figure 4The concentrations of Osa-miR156 in bands 1-15 are 1. 10 fmol; 2. 5 fmol; 3. 2.5 fmol; 4. 1 fmol; 5. 0.5 fmol; 6. 0.25 fmol; 7. 0.1 fmol; 8. 0.05 fmol; 9. 0.025 fmol; 10. 0.01 fmol; 11. 0.005 fmol; 12. 0.0025 fmol; 13. 1 amol;
[0085] From Figure 4 It can be seen that this method can detect at least 0.005 fmol of miRNA (5aM=35fg=0.035pg), that is, band 11 can be clearly identified, and the sensitivity of this method is 10 times higher than that of the previous small RNA detection method.
Embodiment 3
[0087] Small molecule RNA quantitative detection test
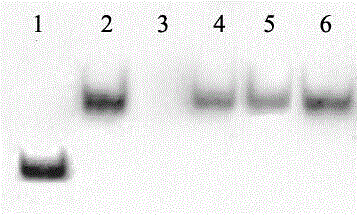
[0088] In this example, the quantification of miR156 in rice was taken as an example; the DNA form miD156 of miR156 with a series of concentration gradients (0.8-31.2 fmol) was hybridized with 0.1 pmol of biotin-labeled probe miD156*, and ChemiDoc XRS was used after color development The detection system carried out relative quantitative analysis. The concentration of miD156 used for hybridization was used as the X axis, and the chromogenic concentration was used as the Y axis to establish a coordinate curve. The results showed a good linear relationship between the two, y = 34.671x + 0.231, and R = 0.9902 in the linear regression equation. Band 7 is the hybridization of 1ug rice miRNA with 0.1pmol miD156* probe, the result is in the regression equation,
[0089] The result is as Figure 5 as shown, Figure 5 In , the miR156 concentrations of bands 1-7 were 1: 0.8fmol; 2:1.9fmol; 3: 3.9fmol; 4: 7.8fmol; 5: 15.6fmol; 6: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com