Digital PCR quantitative detecting kit for HBV (hepatitis B virus) cccDNA and application of digital PCR quantitative detecting kit for HBV cccDNA
A quantitative detection and kit technology, applied in the field of in vitro nucleic acid detection of viruses, to achieve the effects of ingenious detection design, high sensitivity and rapid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: Preparation of DNA in Liver Puncture Tissue
[0047] Liver tissue puncture was performed according to medical routine. Take the liver tissue obtained by 5mm puncture, cut it into pieces with ophthalmic scissors, wash with 200 μL of PBS buffer, and centrifuge at 800 rpm to remove the supernatant, a total of 3 times. Genomic DNA was extracted from the washed liver tissue using the QIAGEN genome mini-extraction kit, and the eluent was 96 μL.
[0048] The extract uses PSAD enzyme to digest HBV rcDNA, the system is as follows:
[0049] DNA extract 32μl
[0050] PSAD enzyme (10U / μl) 2.0 μl
[0051] 10× Digestion Buffer 4.0 μl
[0052] ATP (25mM) 2.0 μl
[0053] The total volume was 40 microliters and incubated for 60 minutes at 37°C followed by 10 minutes at 95°C.
Embodiment 2
[0054] Embodiment 2: Design and synthesis of HBV cccDNA and RPP40 specific primer probe
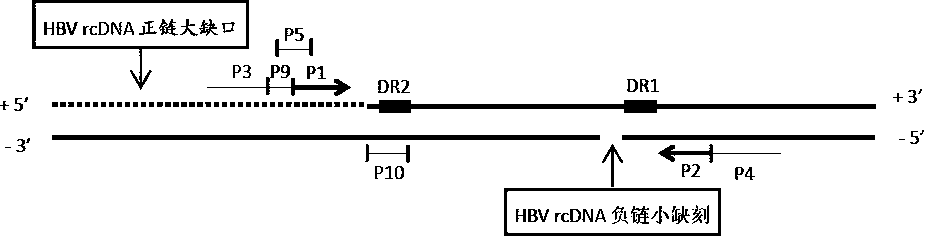
[0055] In the upstream of the DR2 region of HBV DNA, the positive strand gap region of HBV rcDNA, according to the conservation of different genotypes of HBV, the HBV-specific primer fragment P1 was designed, and a random sequence P9 (SEQ ID No. 9) was linked to its 5' end, The nucleotide sequence shown in SEQ ID No. 1 is formed after the common upstream primer P3 (SEQ ID No.3) is linked upstream of P9, and this nucleotide sequence is the HBV cccDNA specific upstream primer. The 11 nucleotide sequences at the 5' end of P1 and the 9 nucleotides at the 3' end of P9 form the HBV cccDNA-specific detection probe P5 (SEQ ID No.5), whose 5' end is labeled with FAM fluorescein, 3' end labeled MGB. In the upstream of the HBV rcDNA minus-strand nick, according to the conservation of different genotypes of HBV, the HBV-specific primer fragment P2 was designed, and its 5' end was linked with the c...
Embodiment 3
[0058] Example 3: Quantitative detection of HBV cccDNA
[0059] (1) PCR reaction system:
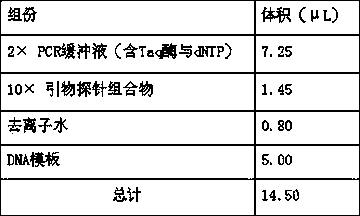
[0060] Prepare the digital PCR reaction system as shown in Table 3;
[0061] Table 3 HBV cccDNA digital PCR quantitative detection reaction system
[0062]
[0063] (2) Detection chip loading:
[0064] According to the requirements of the digital PCR user manual, load the prepared PCR reaction solution into the reaction chip through the chip loading hole, and then seal the reaction chip with 5.50 μL blocking solution;
[0065] (3) PCR reaction conditions:
[0066] Carry out PCR amplification by the reaction conditions shown in table 4;
[0067] Table 4 Reaction conditions for quantitative detection of HBV cccDNA digital PCR
[0068]
[0069] (4) Interpretation of digital PCR results:
[0070] Quantification of total HBV cccDNA in the reaction system = number of blue fluorescent microwells;
[0071] Quantification of the total cell number in the reaction system = 0.5×number o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com