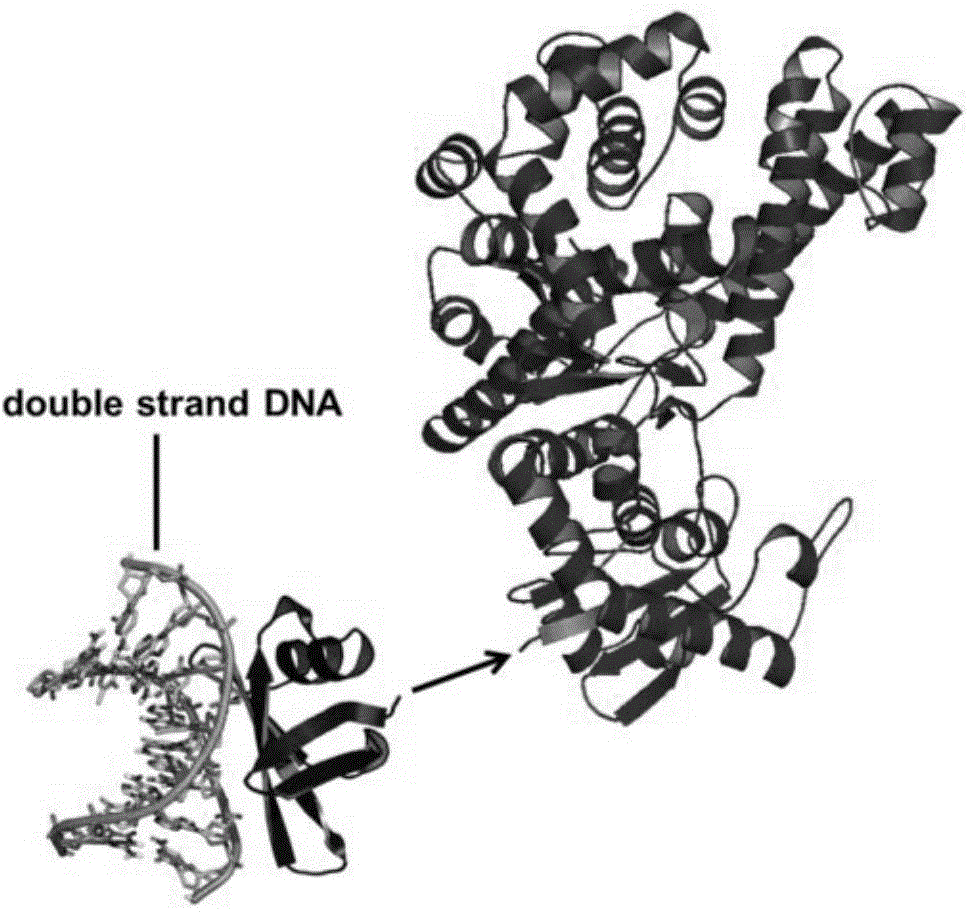
Sso7d-Sau recombinant DNA polymerase
A polymerase and N-terminal technology, applied in the fields of molecular biology and protein engineering, can solve the problem of low continuous synthesis ability of DNA polymerase, achieve high tolerance, wide application prospects, and improve the effect of DNA amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] This example is used to illustrate the acquisition of Sso7d-Sau fragments.
[0036] 1. Synthesize the Sso7d-linker sequence as shown in SEQ ID No.3, wherein the Sso7d sequence refers to the sequence information published in GenBank (NCBI reference sequence number is NC_002754.1), and the linker sequence is GGAGGCGGAGGCTCA GGAGGCGGAGGCTCA GGAGGCGGAGGCTCA.
[0037] 2. Design and synthesize primers for Sso7d-linker sequence and Sau gene sequence published by GenBank (NCBI reference sequence number is YP_006237943.1):
[0038] Sso_F: 5'-CATG CCATGG CAACAGTAAAGTTCAA-3'(NcoI) (SEQ ID No.4)
[0039] Linker_R: 5'-GTCGACGCTTGCTGATGAGCCTCCG-3' (SEQ ID No.5)
[0040] PS_F: 5'-CGGAGGCTCATCAGCAAGCGTTG-3' (SEQ ID No.6)
[0041] PS_R: 5'-TTTTCCTTTT GCGGCCGC TTTTGCATCATACCAAGTTGC-3'(NotI) (SEQ ID No.7)
[0042] 3. Use specific primers Sso_F / Linker_R to use the artificially synthesized plasmid pUC57-Sso7d-linker containing the Sso7d-linker gene as a template for PCR amplification; ...
Embodiment 2
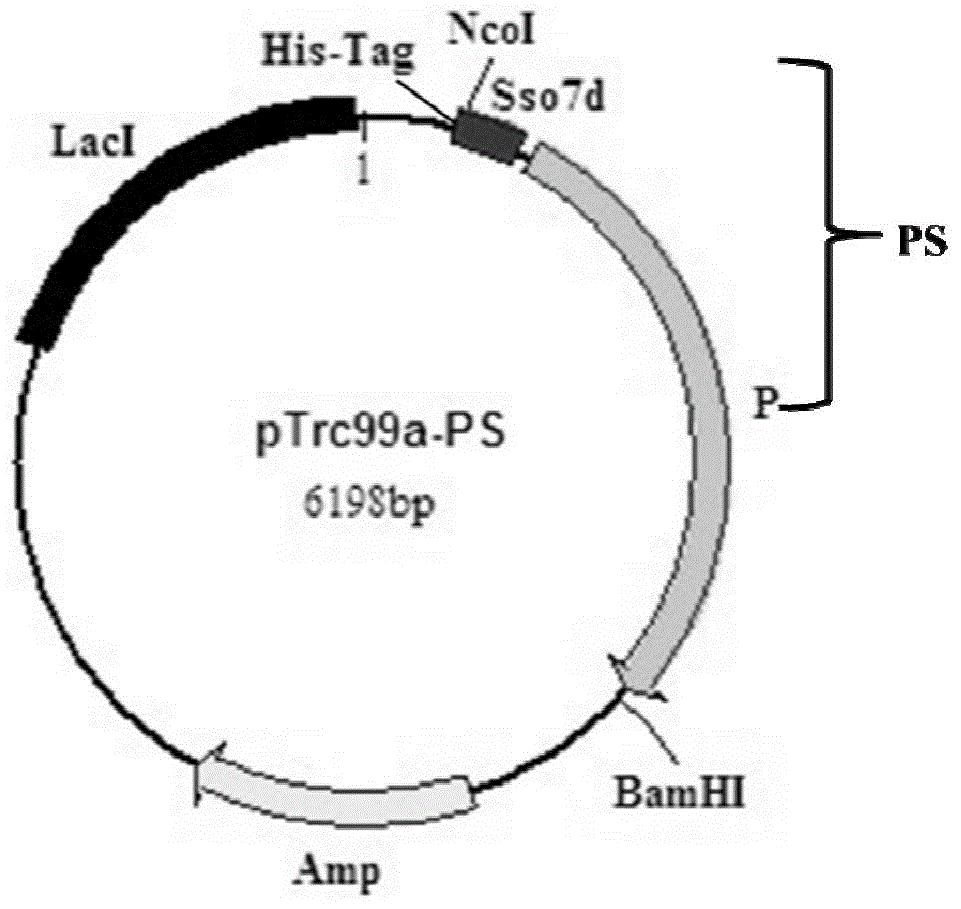
[0057] This embodiment is used to illustrate the construction of recombinant plasmid pTrc99A-Sso7d-Sau ( figure 2 ).
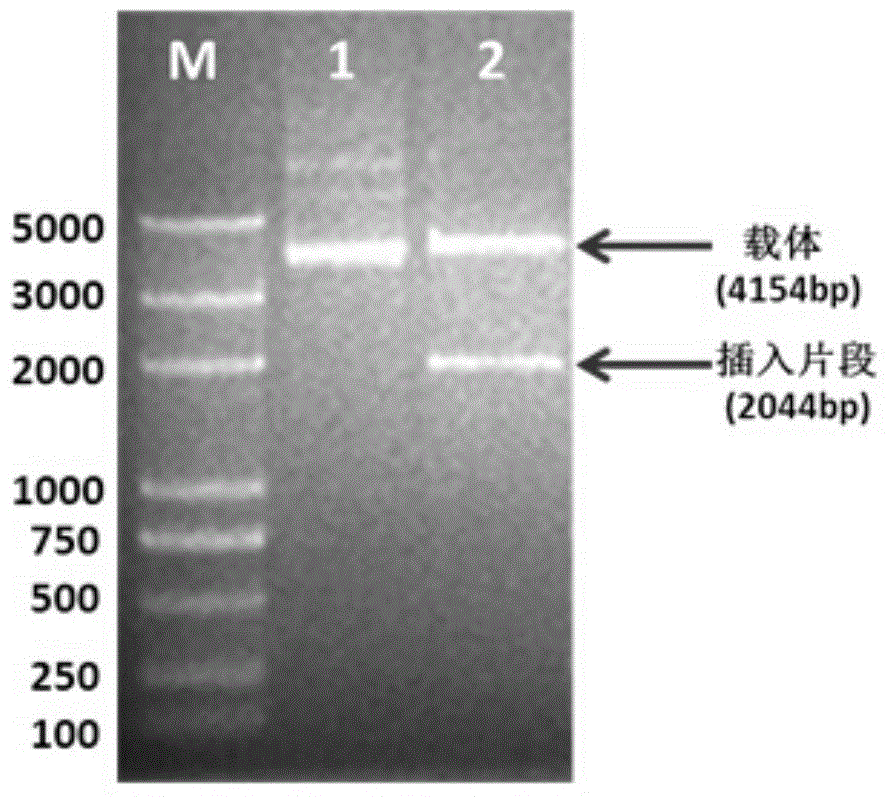
[0058] 1. Sso7d-Sau fragment and pTrc99A vector were digested at 37°C for 2 hours, and the digested products were recovered by gel. The enzyme digestion system is as follows:
[0059]
[0060] 2. Ligate the digested Sso7d-Sau and pTrc99A fragments with T4 ligase, and ligate overnight at 16°C. The connection system is as follows:
[0061]
[0062] 3. Transformation: Take out the Escherichia coli DH5α competent cells from the -70°C refrigerator and bury them in ice cubes, take out the ligation product from the 16°C water bath, remove the parafilm, and put them on ice. After the competent cells melt, put Add all the ligation products to 100 μl DH5α competent cells and mix well. On ice for 30 minutes. Heat shock at 42°C for 90s, and act on ice for 2min. Then add 800 μl LB, shake at 180 r / min for 1 hour at 37°C. The culture was centrifuged at 5000r / mi...
Embodiment 4
[0082] This example illustrates the induced expression and purification of recombinant proteins.
[0083] 1. Transform the positive recombinant expression plasmid pTrc99A-Sso7d-Sau verified by sequencing into Escherichia coli expression strain BL21(DE3), pick a single colony and culture in LB liquid medium containing kanamycin overnight at 37°C .
[0084] 2. Inoculate the overnight bacterial solution into 500ml LB liquid medium containing kanamycin at a ratio of 1 / 100, and culture it on a shaker at 37°C until OD 600 When the value reached 0.6, IPTG was added to a final concentration of 0.5 mmol / L for induction. At the same time, the expression vector control group was set, and the bacterial liquid was taken 4 hours after induction of expression, centrifuged at 12 000 r / min for 10 minutes, and the supernatant was removed.
[0085] 3. Ni-NTA sample buffer (20mM Na 3 PO 4 , 0.5M NaCl, 10mM imidazole, pH 7.4) to resuspend the bacterial pellet, after ultrasonic disruption, cent...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com