A method for establishing a Lactobacillus reuteri non-resistance marker gene integration system and its application
A technology of Lactobacillus reuteri without resistance markers, which is applied in the field of genetic engineering to achieve the effect of avoiding mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Cloning of pheS gene and construction of its mutant gene
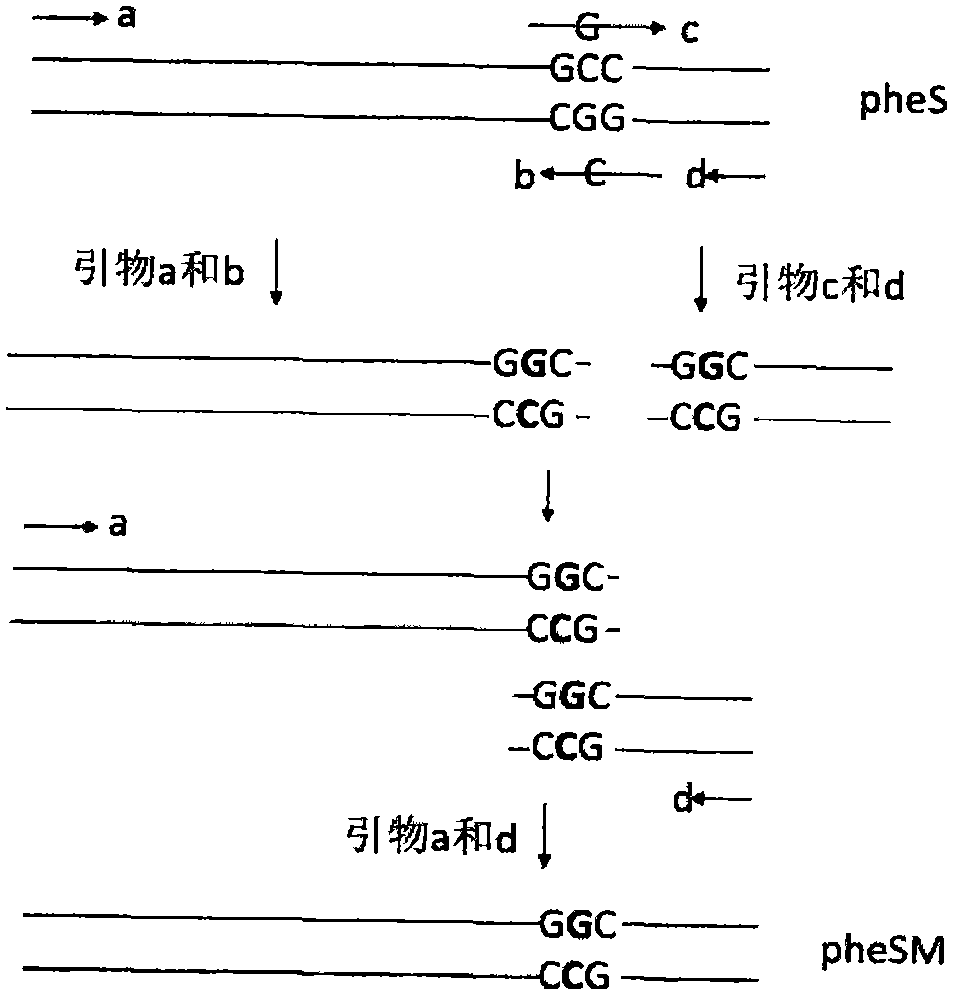
[0046] Design specific primers according to the Lactobacillus reuteri pheS gene sequence published in Genebank, then use the Lactobacillus reuteri XNY genomic DNA as a template to amplify, connect to the cloning vector pMD19-T for sequencing, and obtain the correct L. reuteri XNY pheS gene; then carry out pheS gene amino acid sequence alignment ( figure 1 ), combined with the published literature to determine the mutation site as the second base (C mutation to G) of the 312th amino acid (Ala), and then use the overlapping PCR technique to construct the point mutation gene pheSM of the pheS gene ( figure 2 ).
Embodiment 2
[0047] Example 2 Construction of pheSM gene recombinant L. reuteri XNYM strain
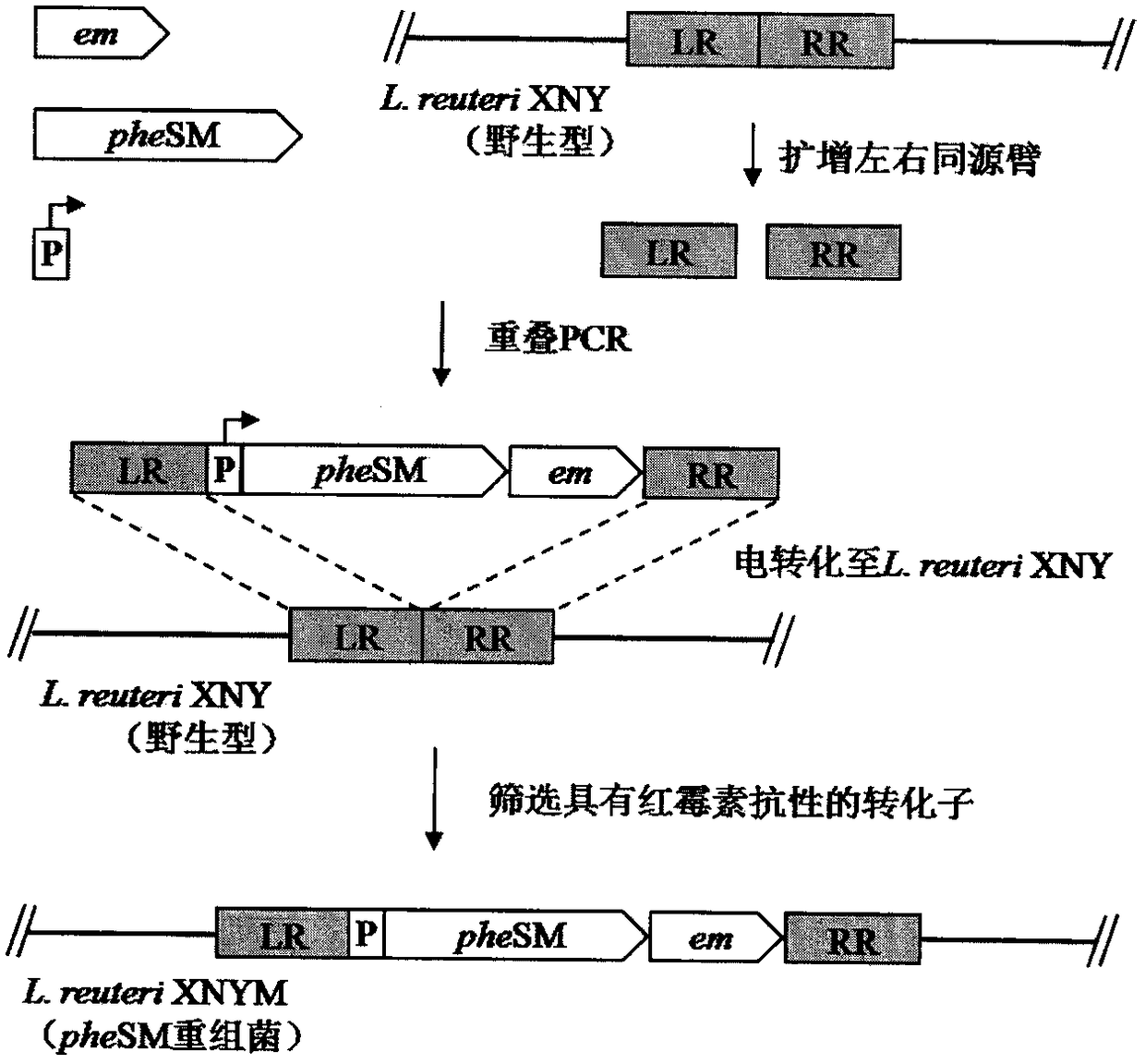
[0048] Select the insertion site of the target gene, and use Lactobacillus reuteri XNY genomic DNA as a template for PCR amplification to obtain 700bp homology arm fragments (LR, RR) upstream and downstream of the insertion site; then amplify the promoter fragment (P), The erythromycin resistance gene (EM) fragment was connected with the pheSM gene and the homology arm fragment by overlapping PCR technology to obtain the LR-P-pheSM-EM-RR integration frame, which was connected to the cloning vector pMD19-T for sequencing, After obtaining the correct transformant, double enzyme digestion to obtain the LR-P-pheSM-EM-RR integration box, electrotransform into L. reuteri XNY competent cells, spread the MRS plate containing erythromycin, and extract the genomic DNA of the positive transformant And carry out PCR detection, obtain pheSM gene recombinant Lactobacillus L.reuteri XNYM ( image 3 ).
Embodiment 3
[0049] Example 3 Construction of L.reuteri recombinant cellulase genetically engineered bacteria
[0050] The cellulase gene cel15 (including the promoter) was connected with the homology arm sequences at both ends of the insertion site by overlapping PCR technology to obtain the cel15 gene integration framework LR-cel15-RR, which was connected to the cloning vector pMD19-T for sequencing to obtain the correct After the transformant, the integrated frame was obtained by double enzyme digestion, electrotransformed into pheSM gene recombinant Lactobacillus L. reuteri XNYM competent cells, spread on the MRS plate containing p-Cl-Phe, extracted positive transformant genomic DNA, and detected by PCR The L. reuteri recombinant bacteria ( Figure 4 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com