Mutants of highly active tumor necrosis factor-related apoptosis-inducing ligands
A tumor necrosis factor and apoptosis-inducing ligand technology, which can be used in anti-tumor drugs, receptors/cell surface antigens/cell surface determinants, pharmaceutical formulations, etc., and can solve problems such as low binding force
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Design of DR5 high-affinity TRAIL mutants
[0031] Construction of the TRAIL-R complex model: the TRAIL-DR5 complex (PDB ID: 1D4V) was used as a template, which is a monomeric complex of TRAIL and DR5 extracellular domain, with a resolution of Wherein DR5 includes amino acids 69-184, and the structure of the TRAIL-DR5 trimer complex was generated using the PDBePISA server (http: / / pqs.ebi.ac.uk) of the EBI website. The structure of the TARIL-DR5 trimer complex obtained in this way must have some unreasonable energy collisions, so the MOE (Molecular Operating Environment, Chemical Computing Group, Montreal) software was used to optimize the energy of the pair: first, the system was added to the system with Protonate 3D Hydrogen, charge, and then under the OPLS-AA force field, set the Gradient to 0.05, and perform energy minimization. The optimized structure is used as the initial structure for subsequent FoldX (http: / / foldxsuite.crg.eu / , Center for Genomic Reg...
Embodiment 2
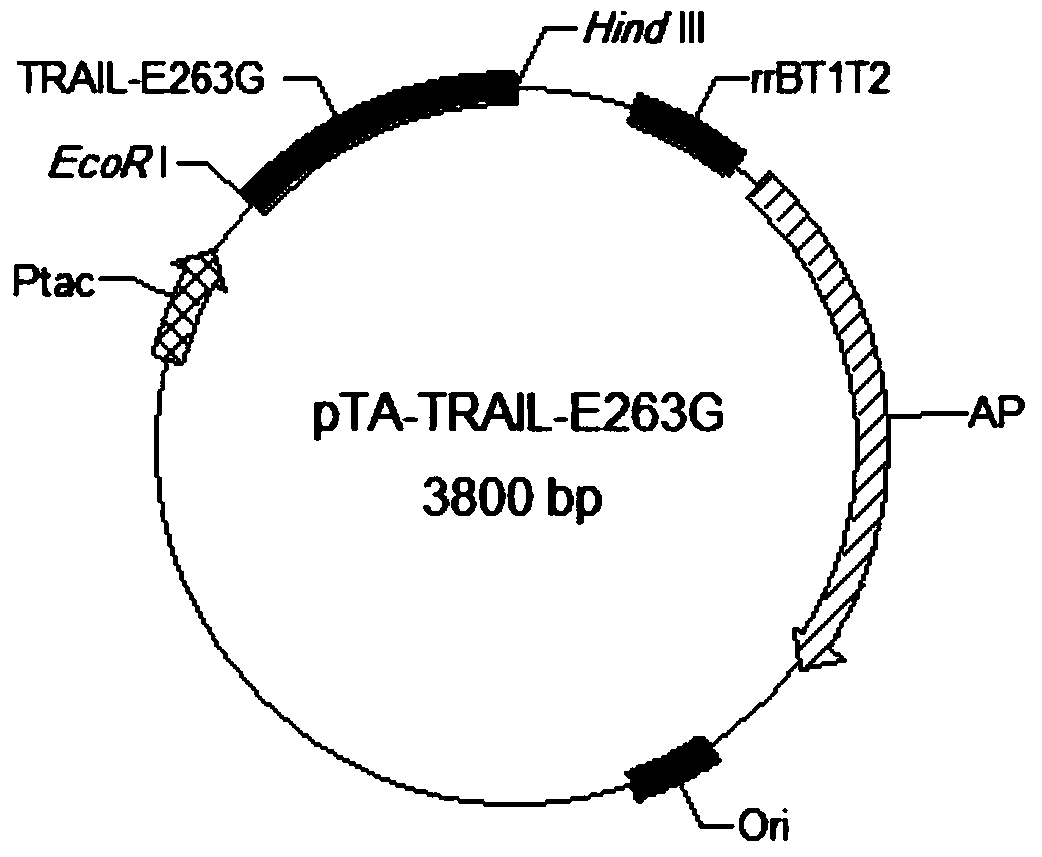
[0040] Example 2 Construction of TRAIL (114-281) and its mutant protein engineering bacteria
[0041] Construction of TRAIL(114-281) expression strain. Design and synthesize the following primers: P1(5-CC GAATTC ATGGTGAGAGAAAGAGGTCCTCAGAGAG-3, SEQ ID NO: 6, the underline is the introduced EcoR I restriction site) and reverse primer P2 (5-AC AAGCTT AGCCAACTAAAAAGGCCCCGAAAAAACTG-3, SEQ ID NO: 7, the underline is the introduced Hind III restriction site). Human placenta cDNA was used as template, P1 and P2 were used as upstream and downstream primers, respectively, and high-fidelity TaqDAN polymerase PCR was used to amplify the TRAIL (114-281) coding gene. PCR conditions were: denaturation at 94°C for 5 minutes; 30 cycles at 94°C for 30s, 50°C for 50s, and 72°C for 90s; extension at 72°C for 10 minutes. After purification, the PCR product was cloned into the pMD19-T vector (product of Dalian Biotech Co., Ltd.) and sent to Shanghai Meiji Biotechnology Co., Ltd. for sequencing...
Embodiment 3
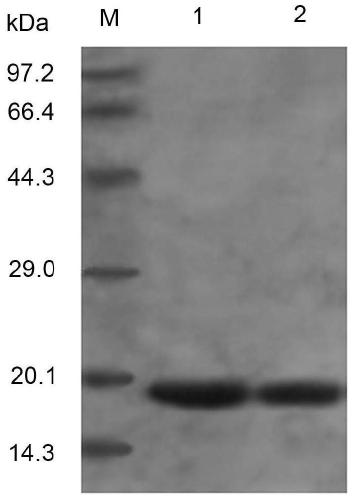
[0043] Example 3 Expression of TRAIL(114-281) and its mutant protein
[0044] A single colony of the recombinant engineered bacteria was placed in 50 ml LB liquid medium (containing 100 μg / ml ampicillin) at 37° C., 220 rpm, and cultured for 12 hours. Inoculate 200ml fermentation medium (1% tryptone, 0.5% yeast powder, 4% sodium glutamate, 1% malt powder, 0.67% KH 2 PO 4 , 0.75% Na 2 HPO 4 . 12H 2 O, 0.1 mM ZnSO 4 , 100 μg / ml ampicillin, pH 6.5), 37°C, 220rpm, cultured for 24h.
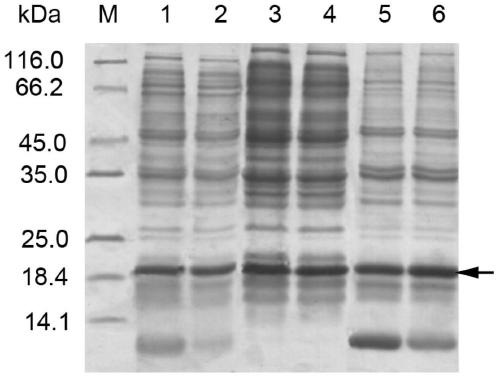
[0045] The fermentation broth was centrifuged (6000rpm, 10min) to collect the bacteria sludge (wet bacteria weight, 6g / l), and resuspended in the bacteriostasis buffer (20mM Tris-HCl, 0.05mM ZnSO 4, 0.25mM DTT, 0.1% Triton, pH 7.6), the cells were disrupted with a homogenizer (AH100B, ATS Engineering Inc., Canada). The broken liquid was centrifuged (13000rpm, 30min) to collect the supernatant. 10% SDS-PAGE electrophoresis analysis of the total protein of the bacteria, the supernatant of the br...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com