Engineering yeast strain for preventing porcine circovirus disease
A technology of porcine circovirus disease and genetically engineered bacteria, applied in the field of genetic engineering, can solve the problems of unfavorable immunization, short residence time of recombinant strains, and high production costs, so as to improve immunogenicity and immune efficacy, and improve non-specific immunity , Improve the effect of specific immunity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
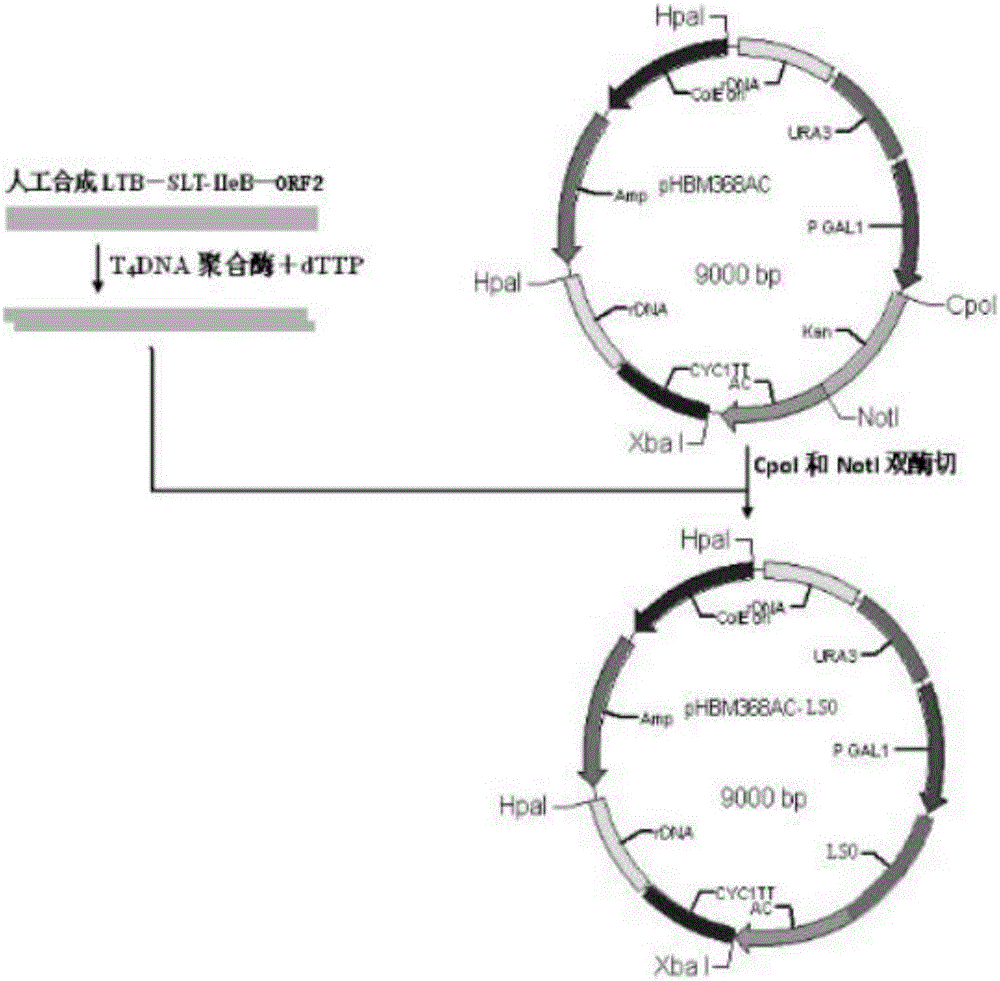
[0042] Embodiment 1 shows the construction of the yeast engineering strain of preventing porcine circovirus disease of antigen LTB, SLT-IIeB and ORF2 fusion protein
[0043] 1) According to the yeast codon preference, transform the LTB gene, SLT-IIeB gene in pathogenic porcine Escherichia coli and the ORF2 of porcine circovirus type 2 virus-encoded viral nucleocapsid protein Cap; design and synthesize the recombinant gene LSO primers;
[0044] 2) LTB, SLT-IIeB and ORF2 after transformation were amplified by overlappingPCR technology, and the full length of the LSO fusion gene was obtained by connecting the linker sequence; the nucleic acid sequence of LSO is shown in SEQ ID NO: 2;
[0045] 3) Saccharomyces cerevisiae display expression vector pHBM368AC was treated with CpoI and NotI double enzymes;
[0046] 4) GTCA was introduced at the 5' end of the fusion gene LSO, and two nucleotide sequences of GGCCA were introduced at the 3' end; the amplified product was treated with T4...
Embodiment 2
[0049] Embodiment 2: the acquisition of S-SP3 recombinant strain
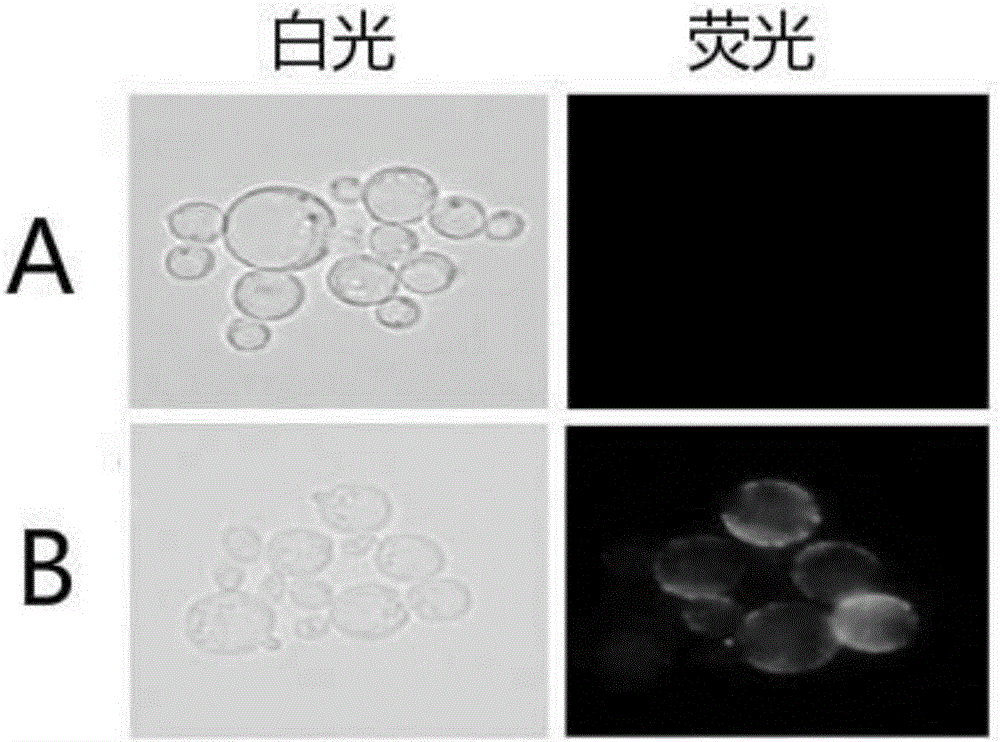
[0050] The recombinant transformants obtained in Example 1 were extracted from the genome, and the positive clones that had been correctly inserted into the LSO gene were identified by PCR; the correct recombinant transformants verified by inoculation were cultured in 50ml YNB-HLAD liquid medium, 28°C, 200rpm for 12-16 hours, then collect the bacteria and transfer them to 100ml YNB-HLAG liquid medium, culture at 28°C, 200rpm for 48 hours, and then centrifuge at 5000rpm for 5 minutes at 4°C to collect the bacteria. The collected bacteria were then subjected to immunofluorescence detection and westernblotting detection, the results are shown in figure 2 .
[0051] figure 2 It is the indirect immunofluorescence detection of the recombinant brewer's yeast INVScl / pHBM368AC-LSO (named S-SP3) of the present invention; where A is the blank control brewer's yeast, and B is the recombinant brewer's yeast, respectivel...
Embodiment 3
[0052] The biological characteristic of embodiment 3S-SP3 recombinant bacterial strain
[0053] 1. Analysis of growth characteristics of S-SP3 recombinant strain
[0054] Streak culture of S-SP3 recombinant strain on solid YEPD plate, pick a single colony into liquid YNB-HLAD medium, culture at 28C, 200r / min for 16 hours, insert 3% inoculum amount into the yeast culture that has been sterilized before Continue to culture in the medium, the temperature of the medium is 28°C, the pH is 4.6, and the rotation speed is 170r / min. Experiments have proved that the genetic engineering strain of the present invention can be fermented at a high density, with an OD600 of over 40, no inducer needs to be added during the whole fermentation process, and it can be strictly expressed at the end of the logarithm. Finally, the biomass of Saccharomyces cerevisiae reached 8.124g / L.
[0055] 2. Genetic stability of S-SP3 recombinant strain
[0056] Streak culture the S-SP3 recombinant strain pre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com