Municipal refuse treatment method
A treatment method, the technology of municipal solid waste, applied in the field of municipal solid waste treatment, can solve the problems that are not suitable for the treatment of large-scale municipal solid waste, achieve high application and promotion value, improve capacity and efficiency, and improve the effect of degradation ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Cloning of embodiment 1 lipase gene
[0024] Using Pseudomonas DNA as a template, use primer 1 to add the EcoRI restriction site:
[0025] GGATCCatgggtgtgtatgactacaaga, with primer 2 added SpeI restriction site:
[0026] ACTAGTtcaggcgatcacgattccatcagc; PCR amplified. The PCR reaction conditions of the gene were: 94°C for 5min, 94°C for 45s, 54.6°C for 40s, 72°C for 103s, 30 cycles, 70°C for 10min, 4°C for eternity.
[0027] A band of about 1854 bp was recovered by the gel recovery system, and its sequence was shown in SEQ ID NO: 1 by sequencing. Through blast comparison in NCBI, it is found that it is a kind of lipase.
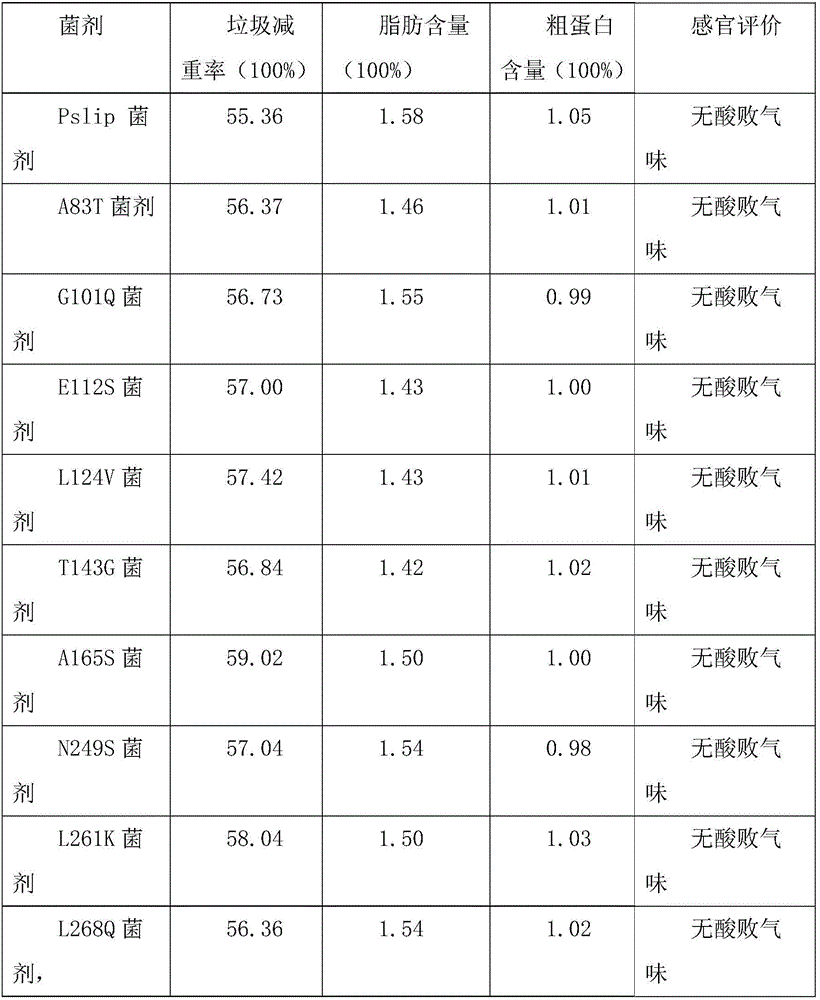
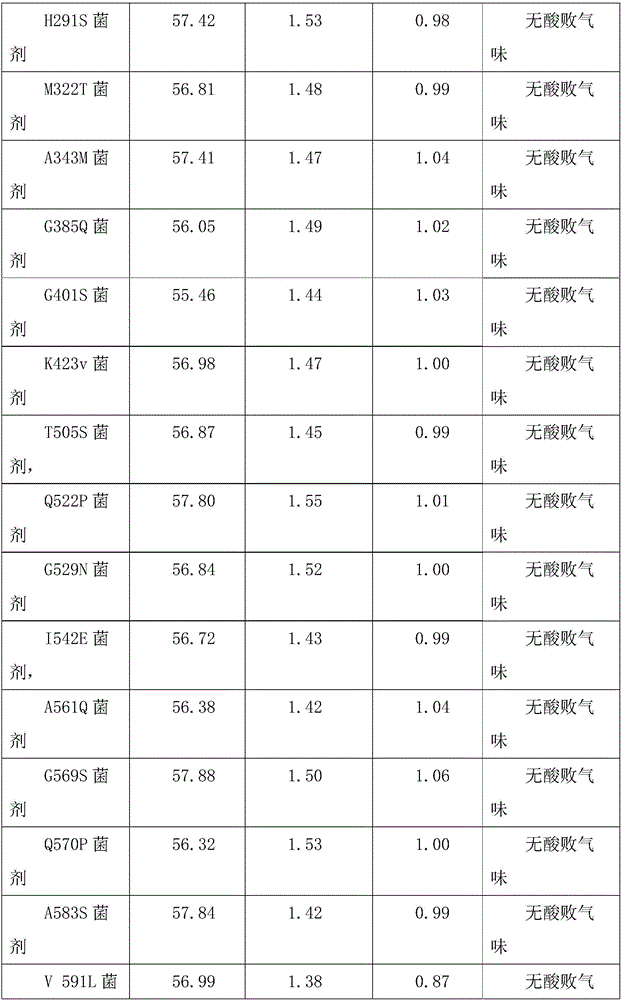
[0028] According to the conventional gene mutation method in this field, the genes were respectively placed in A 83T, G 101Q, E112S, L124V, T 143G, A 165S, N 249S, L 261K, L 268Q, H 291S, M 322T, A 343M, G 385Q, G 401S, K423v, T 505S, Q 522P, G 529N, I 542E, A 561Q, G 569S, Q 570P, A583S or V 591L are subjected to point mutations. The specific embod...
Embodiment 2
[0029] The preparation of embodiment 2 transgenic yeast cells
[0030] The coding sequence obtained in Example 1, the corresponding mutant sequence, and the pScIKP vector were double-digested with restriction endonucleases EcoRI and SpeI, respectively, and the products after the double-digestion were purified, and the two were ligated overnight with T4DNA ligase, and transformed into a plate , to identify recombinants, 57 positive recombinants were obtained through PCR identification. Among them, the recombinant plasmid pScIKP-Pslip was successfully obtained through plasmid extraction and identification. At the same time, similar recombinant plasmids pScIKP-Pslip-A 83T, pScIKP-Pslip-G 101Q, pScIKP-Pslip-E 112S, pScIKP-Pslip-L 124V, pScIKP-Pslip-T143G, pScIKP-Pslip-A 165S, pScIKP-Pslip-N 249S, pScIKP-Pslip-L 261K, pScIKP-Pslip-L 268Q, pScIKP-Pslip-H 291S, pScIKP-Pslip-M 322T, pScIKP-Pslip-A 343M, pScIKP-Pslip-G 385Q, pScIKP -Pslip-G 401S, pScIKP-Pslip-K 423v, pScIKP-Pslip-T50...
Embodiment 3
[0034] The preparation of embodiment 3 bacterial agents
[0035] Alkaline xylosoxidans denitrifying subspecies CGMCC 1.768, Bacillus amyloliquefaciens CGMCC 1.1177, Debaryomyces hansenii var. Nocardia pasteuri CGMCC 4.1128, each bacteria was expanded and cultivated according to the optimal medium, and the volume ratio of the mixed bacteria solution was 10:5:10:1:5:10 to prepare the corresponding bacterial agent. The concentration of each bacteria before mixing is 2*108 / ml.
[0036] The lipase gene or its variants were introduced into Debaria hansenii var. hansenii CGMCC 2.1831 and Issa rhomboides CGMCC 2.1592 respectively to replace the original Debariea hansenii var. hansenii CGMCC 2.1831 without lipase gene , Issahia rhizoma rhomboides CGMCC 2.1592 corresponding to the preparation to become the corresponding bacterial agent. Bacteria named: Pslip Bacteria, A83T Bacteria, G101Q Bacteria, E112S Bacteria, L124V Bacteria, T143G Bacteria, A165S Bacteria, N249S Bacteria, L261K B...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com