Soybean type bacillus subtilis proteinase gene and application
A technology of Bacillus subtilis and protease, applied in the field of molecular biology, can solve problems such as unreported
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] The clone of the cDNA sequence of embodiment 1 gene Gmsbt1.6-3
[0030] The soybean (Glycine max) variety Jinong 18 and the soybean root mutant material M18 were provided by the Plant Biotechnology Center of Jilin Agricultural University.
[0031] Cloning of cDNA sequence of gene Gmsbt1.6-3
[0032] Total RNA was extracted from the root tissue of soybean M18 seedlings using the RNAiso kit produced by TAKARA, and the reverse-transcribed cDNA was used as a template. According to the sequence of RNA-seq sequencing results, specific primers sbt were designed, and the target fragment was amplified by RT-PCR The PCR amplification primers required for the test are listed in Table 1.
[0033] Table 1 PCR primer sequences
[0034]
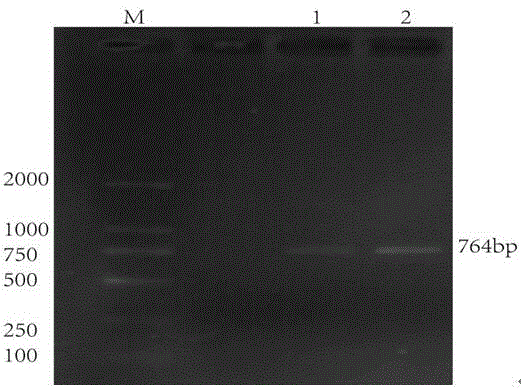
[0035] According to the Gmsbt1.6-3 sequence fragment obtained in the previous screening, a specific primer sbt with an amplification length of 764bp was designed, and after PCR amplification, a nucleic acid band with the same size as expected wa...
Embodiment 2
[0036] Example 2 Gmsbt1.6-3 Gene bioinformatics analysis and phylogenetic tree construction
[0037] Will Gmsbt1.6-3 The full sequence of the gene was connected into the cloning vector pMD18-T Vector, and the recombinant vector was sequenced for bioinformatics analysis. The sequencing was completed by Beijing Sanbo Yuanzhi Biotechnology Co., Ltd.
[0038] Use the online analysis program ORF finder (https: / / www.ncbi.nlm.nih.gov / orffinder / ) to analyze the possible open reading frame of the new gene sequence, and translate the new gene ORF into an amino acid sequence through the DNAMAN software; use ProtParam Software (http / / www. web. expasy. org / protparam / ) to predict theoretical parameters of protein sequences; constructed using SWISS-MODEL online software Gmsbt1.6-3 The three-dimensional protein structure of the gene; use Protscale software (http: / / web.expasy.org / / protscale / to predict the hydrophilicity and hydrophobicity of the protein.
[0039] Download all known s...
Embodiment 3
[0044] Construction and genetic transformation of embodiment 3 plant expression vector
[0045] restriction endonuclease Bgl II and Pml I carried out double digestion of the plant expression vector pCAMBIA3301, and utilized the Seamless Assembly Clonng Kit kit to clone the obtained Gmsbt1.6-3 The full-length gene replaces the original one on 3301 GUS Gene. build with bar As a screening marker, the CaMV35S promoter promotes the target gene Gmsbt1.6-3 pCAMBIA3301- Gmsbt1.6-3 Plant overexpression vector.
[0046] use Bgl II and Pml The plant expression vector pCAMBIA3301 was double-digested with I restriction endonuclease to obtain a large fragment of the 3301 vector of 10kb and a GUS gene fragment of 2029bp; the cloning vector Pmd18-T-Gmsbt1.6-3 was carried out by using primer sbt-over PCR amplification, the Gmsbt1.6-3 gene ORF full-length fragment with a size of 591bp was obtained ( Figure 4 ), using T4 ligase to connect the full-length ORF fragment of Gmsbt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com