In-situ hybridization probe and method for identifying barley chromosome set by adopting same
A fluorescence in situ hybridization and chromosome technology, applied in the field of molecular cytogenetics, can solve the problems affecting the test process and effect, complex target sequence process, unsatisfactory labeling effect, etc., to achieve good detection effect, good sensitivity, and easy to distinguish Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
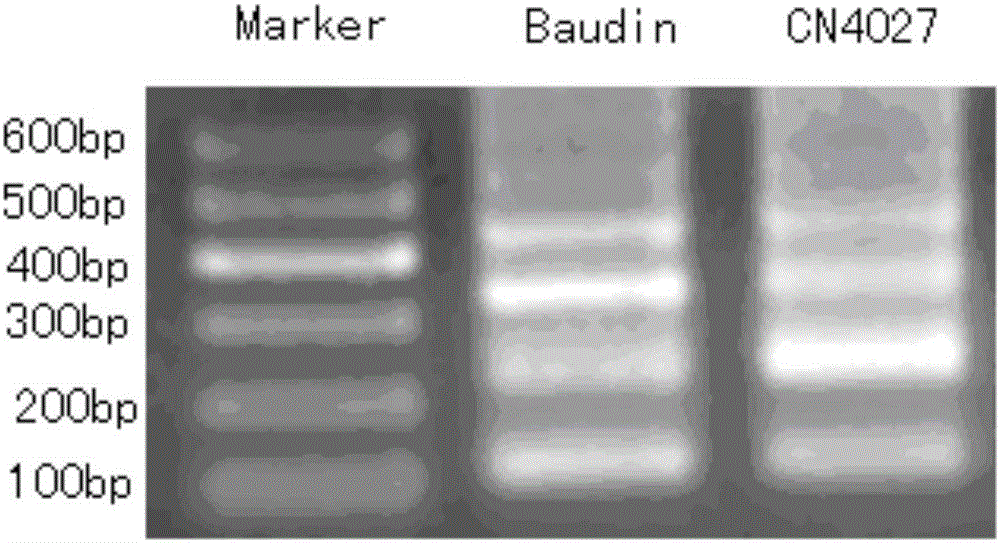
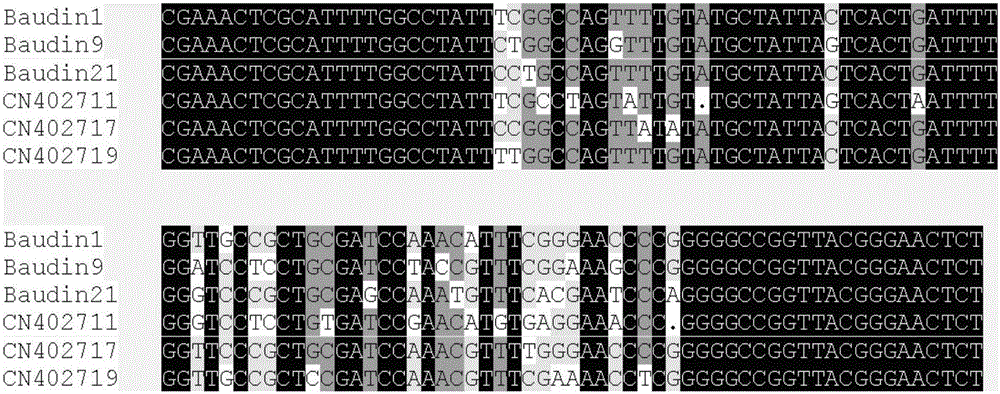
[0036] Example 1 Analysis of Barley Subtelomere Repeat Sequence and Determination of Oligonucleotide Probe Sequence
[0037] 1. Extraction of barley genomic DNA
[0038] Select the young leaves of Baudin and CN4027 to be tested, and use the column method (TaKaRa MiniBEST PlantDNA Extraction Kit) to extract DNA. The extraction steps are as follows:
[0039] a) Take 150 mg of fresh young leaves, grind them into fine powder with liquid nitrogen, quickly add 500 μl of Buffer HS I and 10 μl of 50×DTT Buffer and mix well, then add 10 μl of RNase A (10 mg / ml), shake and mix well, and then Incubate in a 56°C water bath for 10 minutes.
[0040] b) Add 62.5 μl of Buffer KAC and mix thoroughly. Place on ice for 5 minutes and centrifuge at 12,000 rpm for 5 minutes. Take the supernatant, add the same volume of BufferGB as the supernatant, and mix well.
[0041] c) Place the Spin Column in the Collection Tube, and move the solution to the Spin Column. Add 500 μl of Buffer WA to the Spi...
Embodiment 2
[0061] Example 2 Method of oligonucleotide probe Oligo-442A01 marking barley chromosome in ND-FISH method
[0062] 1. Chromosome preparation of barley root tip cells
[0063] Choose root length 1-2cm root tip after germination in N 2 Treated in O for 4h, inactivated by acetic acid, ddH 2 O wash, cut out the meristematic area, and enzymolyze it in the enzyme mixture (cellulase: pectinase = 1:1) at 37°C for 37 minutes, absorb the enzyme solution and use ddH 2 O and absolute ethanol were washed twice. Add 20 μl of acetic acid to each root tip and stir well until the cells appear in suspension, and drop 10 μl of the suspension on each slide. Examine the chromosomes under an optical microscope, select slides with clear and well-dispersed chromosomes, and mark them and store them in a refrigerator at 4°C for later use.
[0064] 2. Chromosome in situ hybridization
[0065] Take out the slides to dry naturally, mix the hybridization solution at 0.35μl 1OD·ml for each slide -1 Th...
Embodiment 3
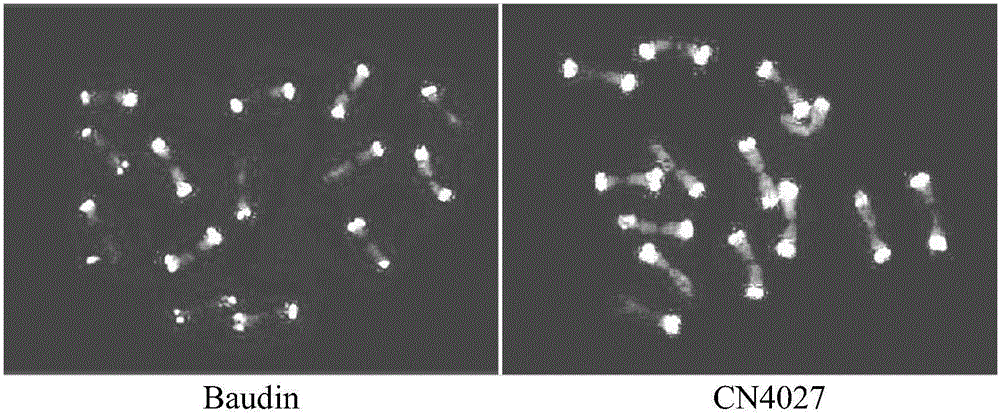
[0067] Example 3 Oligonucleotide probe Oligo-442A01 and (AGG) 5 Establishment and application of barley genome identification method in ND-FISH method
[0068] Select the prepared glass slides in Example 2 to dry naturally, and mix the hybridization solution at 0.35 μl 1OD·ml for each slide -1Oligo-442A01, 0.35μl 1OD·ml -1 (AGG) 5 The ratio of hybridization solution to 9.30 μl was prepared, and the mixed hybridization solution was mixed repeatedly with a pipette gun, and 10 μl of the mixed hybridization solution was pipetted and dropped on a dry glass slide, and the coverslip was incubated in a humidified dark box at 37°C for 2 hours. After taking out, wash 2 times with 2×SSC under dark condition, ddH 2 O wash 1 time, 5min each time. After natural drying, 10 μl DAPI was added dropwise to each slide, and the coverslips were observed under the OLYMPUS BX63 fluorescence microscope for chromosomes and photographs were taken with the OLYMPUS DP80CCD camera. The above hybridiza...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com