Kit used for pig Delta coronavirus detection and detection method thereof
A coronavirus and kit technology, applied in the field of genetic engineering, can solve the problems of long reaction time and low sensitivity, and achieve the effects of high sensitivity, simple operation and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] The design of embodiment 1 specific primer and probe
[0067] Design a pair of specific primers and probes for the conserved sequences of PDCoV in GenBank, as shown in Table 1 below, and synthesize specific fluorescent RT-PCR primers and probes according to the following sequences. with DEPC-H 2 O dilute the primers to 10 μM and store at -20°C for later use.
[0068] Table 1 Primer and Probe Combinations
[0069]
[0070] Wherein the nucleotide sequence of F1 is as shown in SEQ ID NO.1 in the sequence listing, the nucleotide sequence of R1 is as shown in SEQ ID NO.2 in the sequence listing, and the nucleotide sequence of P1 is as shown in SEQ ID NO in the sequence listing .3 shown.
Embodiment 2
[0071] The establishment and optimization of embodiment 2 reaction system
[0072] 1. Sample preparation: construct a plasmid containing the target amplification region as a positive control for PDCoV detection; positive samples of PEDV vaccine strain, TGEV vaccine strain, rotavirus vaccine strain, Jieshen virus, Kobu virus, Escherichia coli ( Standard strain ATCC 25922) (taken from Shanghai Animal Disease Prevention and Control Center Shanghai Veterinary Disease Diagnosis Center Laboratory) as a specific reference; with deionized water as a negative control, Trizol was used to extract the positive control and specific reference respectively RNA, negative control for use.
[0073] 2. Screening of primer probes: detect the RNA of the above-mentioned positive control and negative control respectively with the primer pair probes designed in Example 1, and through repeated trials, screen out the best primer probes with good specificity, sensitivity and repeatability. Needle combi...
Embodiment 3
[0081] The establishment of embodiment 3 standard curve
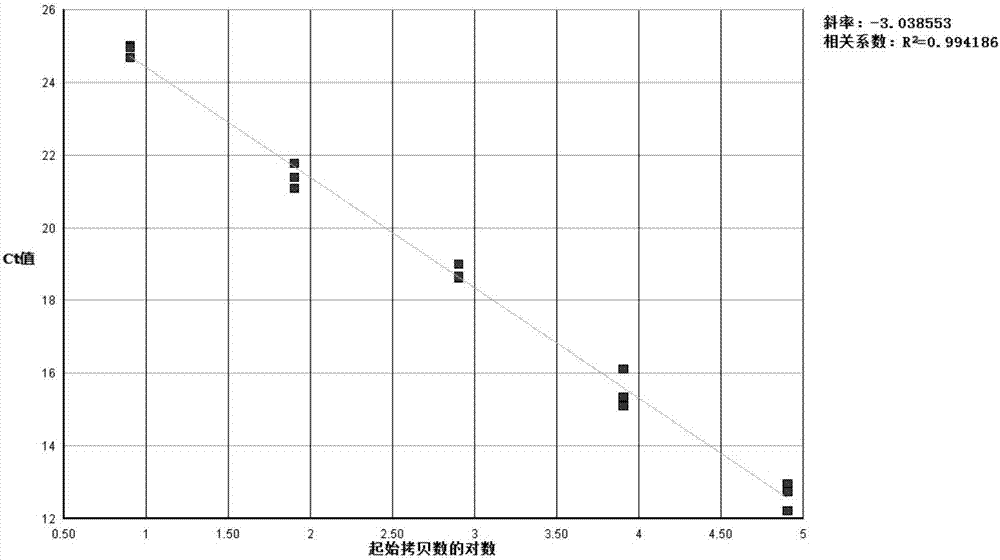
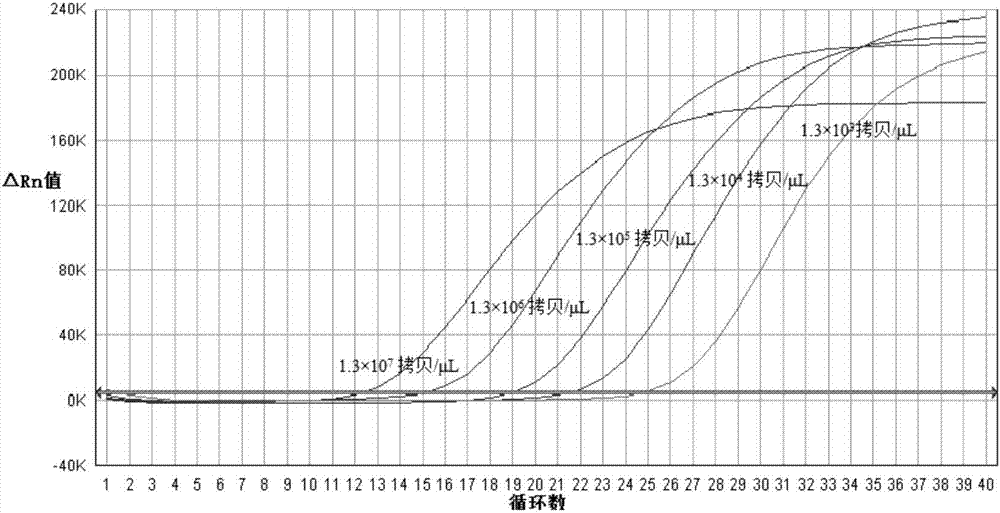
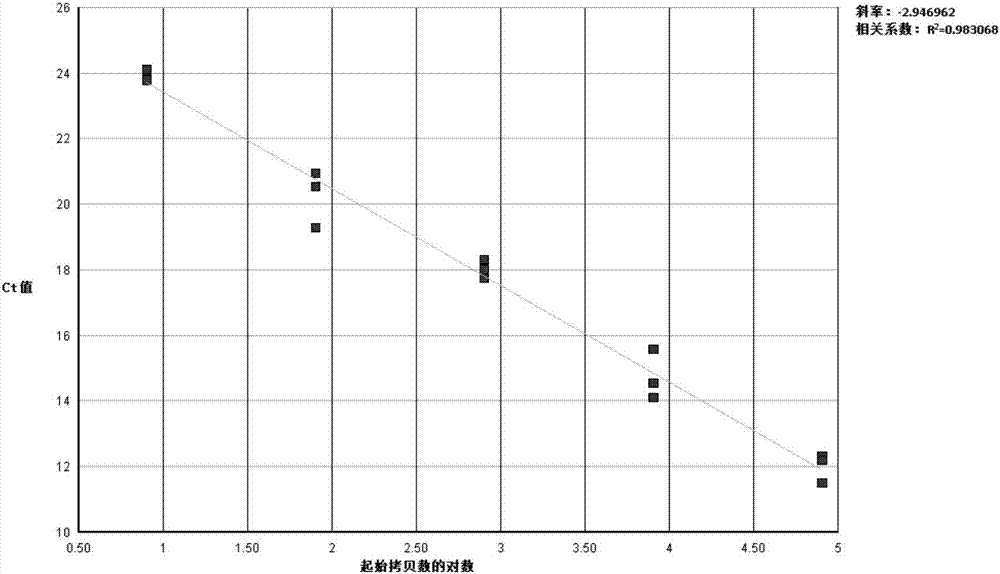
[0082] After measuring the concentration of PDCoV positive plasmid DNA with a spectrophotometer, use DEPC-H 2 O Make a 10-fold serial dilution of the plasmid standard so that the copy number in each 5 μL detection volume is 1.3×10 7 , 1.3×10 6 , 1.3×10 5 , 1.3×10 4 , 1.3×10 3 copy / μL, set 3 repetitions, and perform amplification. After the reaction, use ABI7500 fluorescence quantitative PCR analysis software to automatically obtain the standard curve. The results show:
[0083] The fluorescent RT-PCR method established with combination 1 of primers (F1 and R1) and probe P1 showed a typical S-type amplification curve with obvious exponential region, and the initial template concentration and Ct value of the standard showed a good correlation. Linear range, slope close to -3.04, correlation coefficient r 2 is 0.994 (see figure 1 , 2 ).
[0084] The sample can be accurately quantified according to the Ct value and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com