Novel reaction system for rapidly constructing plant gene site-directed knockout carrier
A site-directed knockout and reaction system technology, applied in the field of molecular biology, can solve the problems that are unfavorable for rapid and high-throughput gene site-directed knockout vector construction, limit vector throughput, and high construction costs, so as to reduce experimental losses and reduce the use of and the effect of reducing the cost of material resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1 Construction method of the vector based on CRISPR-Cas system
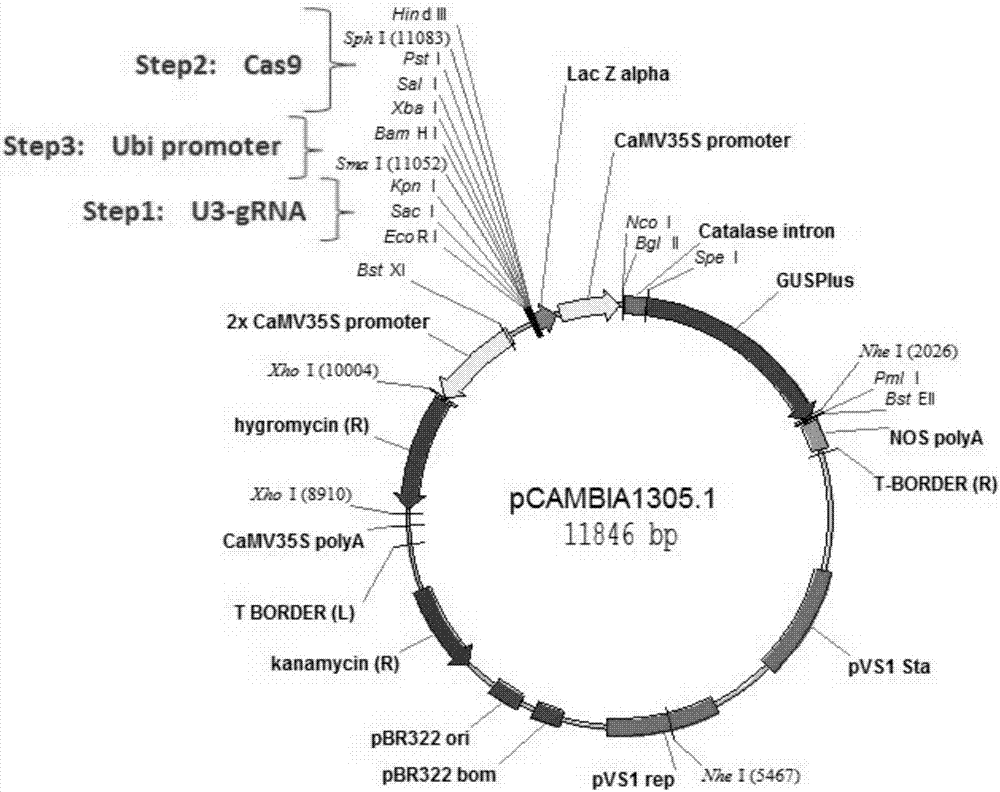
[0048] 1) Use the pOsU3-gRNA plasmid as a template, use sgRNA recombination primers (Table 1) to amplify the OsU3-sgRNA sequence, and homologously integrate it into the pCAMBIA 1305.1 linear vector after double digestion with EcoRI and KpnⅠ
[0049] ( figure 1 ). The ligation product was transformed into DH5a, and positive clones were selected for sequencing. A new plasmid vector containing the correct sequence of OsU3-sgRNA was obtained by sequencing, named pCAMBIA 1305.1-sgRNA;
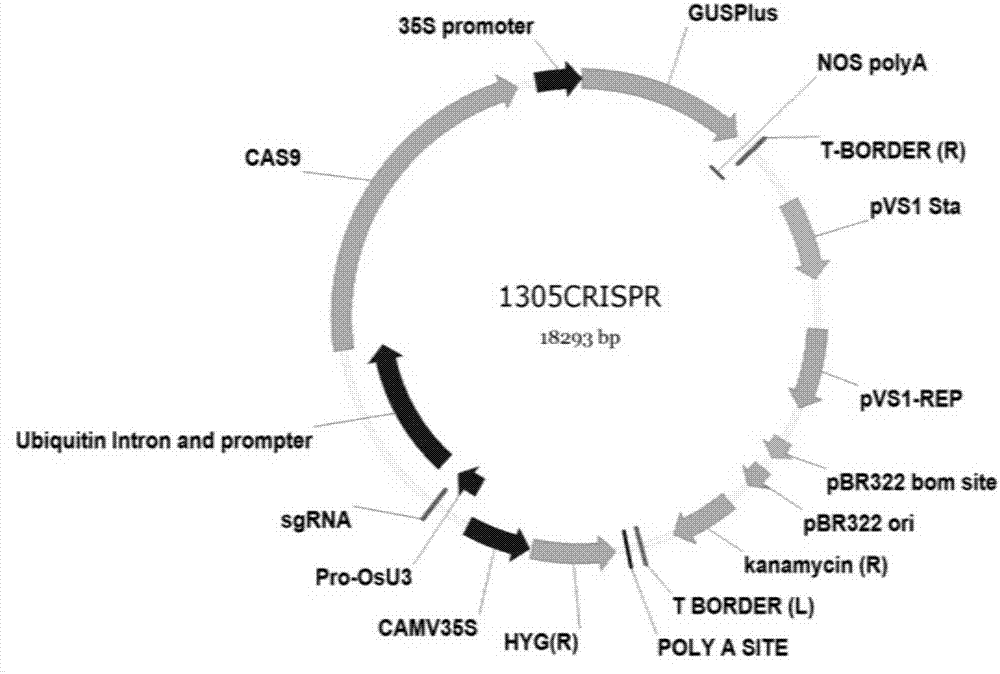
[0050] 2) Using the pJIT163-2NLSCas9 plasmid as a template, the genomic sequence encoding Cas9 was amplified using Cas9 recombination primers (Table 1), and integrated into the pCAMBIA 1305.1-SgRNA vector digested with Xba I and Hind III by homologous recombination. The ligation product was transformed into DH5a, and positive clones were selected for sequencing. A new plasmid containing the correct sequence of the...
Embodiment 2
[0054] Example 2 The reaction system capable of rapidly constructing rice gene-directed knockout vectors
[0055] ① Appropriate amount of 100ng of 1305CRISPR plasmid DNA prepared in 1; ② 10X buffer 1 μL of restriction endonuclease AarⅠ; ③ 0.2 μL of 50X Oligo of restriction endonuclease AarⅠ; ④ 1 μL of double-stranded DNA of target site; ⑤ Restriction endonuclease AarⅠ0. 2 μL; ⑥T4 DNA ligase 0.1 μL; ⑦ATP 1 μL, add water to a total volume of 10 μL.
Embodiment 3
[0056] Example 3 The method for rapidly constructing rice gene-directed knockout vector
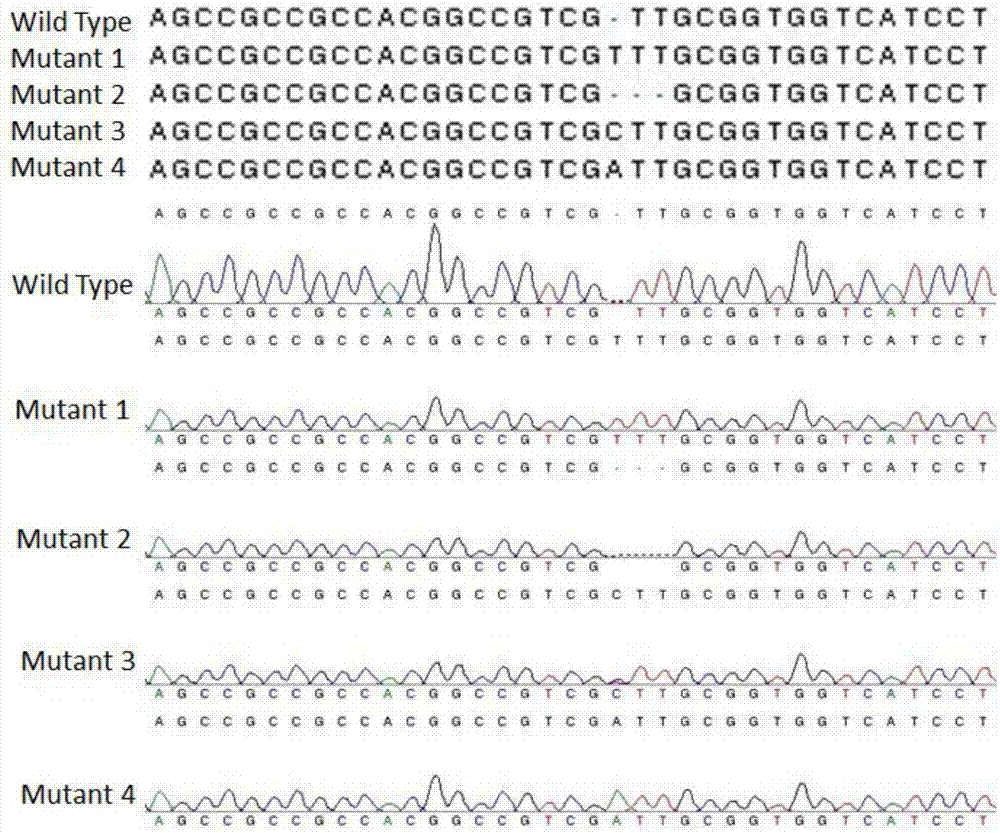
[0057] The reaction system described in Example 2 was placed at 37° C. for 5 minutes and 16° C. for 5 minutes. After 5-10 cycles, the reaction product was directly used to convert DH5a. After the colonies grown on the plate were cultured overnight at 37°C, 1-3 colonies were randomly selected and sent to the company for sequencing confirmation. Any plasmid containing the target gene sequence at the cloning site is the correct plasmid that has been constructed.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com