Recombinant aspergillus flavus chitosanase and encoding gene, as well as preparation and application
A kind of Aspergillus flavus chitosanase and coding gene technology, applied in the fields of application, glycosylase, genetic engineering, etc., can solve the problems that the production and activity of chitosanase are difficult to meet the application, and the activity needs to be improved, so as to achieve the goal of fermentation enzyme Vitality improvement, high-efficiency secretion expression, and high-efficiency degradation effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
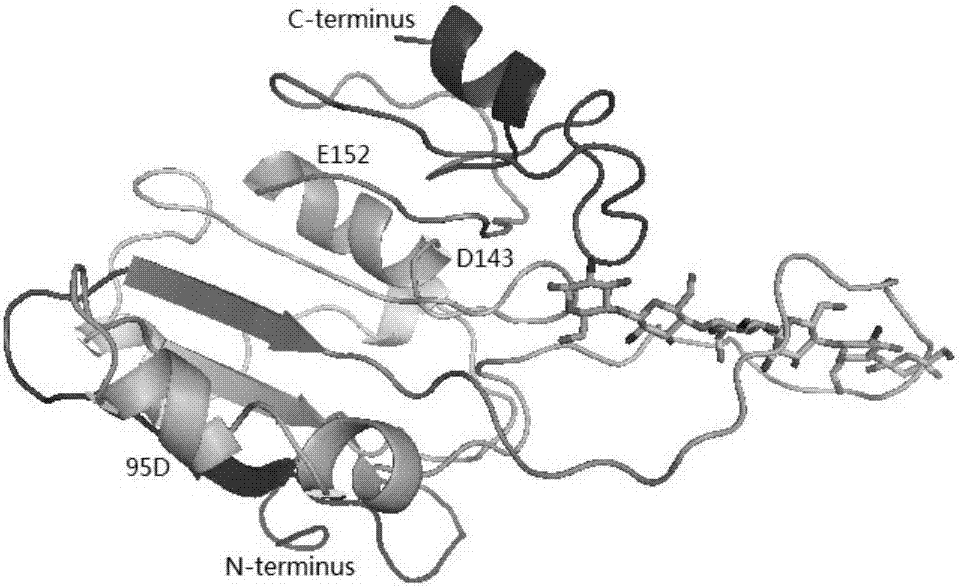
[0024] Example 1 Aspergillus flavus endo-chitosanase 3D model construction
[0025] Using the homologous modeling server Swiss-Model (http: / / swissmodel.expasy.org), the chitosanase mutants are automatically modeled, and the 3D structure is as follows figure 1 . The N-terminal of the chitosanase mutant is composed of 3 sheets and 3 helical sheets, and the C-terminal is composed of 2 sheets and a linear structure, connected by a linker in the middle. from figure 1 It can be seen that the chitosanase mutant position 95D, away from the catalytic active center (D143 / E152), will not affect the catalytic activity of the enzyme
Embodiment 2
[0026] Embodiment 2 mature chitosanase mutant gene cloning
[0027] Taking the Aspergillus flavus chitosanase gene as a reference (the sequence was synthesized by Suzhou Jinweizhi Co., Ltd., GenBank number AY190324, the mature recombinant chitosanase gene was mutated at two sites A286G and C287T, and the corresponding amino acid residue 95D was mutated to 95G ), designed amplification primers, C-F: 5'-TCT CTCGAG TACAACCTACCCAAC-3' and C-R:5'-CTC GCGGCCGC TTAAGCCTTCAAACCAGCAAC-3', wherein the underline is respectively XhoI, NotI enzyme cutting site, and italics is the Kex2 signal cutting site, then carry out polymerase chain reaction (PCR) amplification to obtain recombinant chitosanase mutant gene (without signal Peptide sequence), where the underlines are the XhoI and NotI restriction sites, and the oblique line is the Kex2 signal cleavage site. PCR reaction was carried out with LATaq DNA polymerase. The reaction conditions were: pre-denaturation at 94°C for 2 min, then ...
Embodiment 3
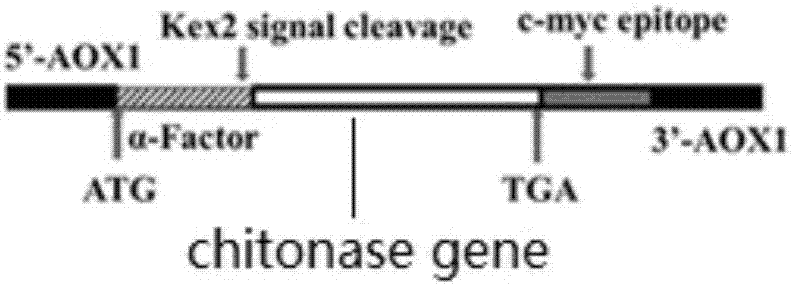
[0051] Embodiment 3 Construction of recombinant plasmid pPICZαA-N-chitosanase mutant
[0052] The construction mode of the recombinant plasmid pPICZαA-N-chitosanase mutant is as follows figure 2 , the α-factor secretion signal sequence gene on the vector pPICZαA is directly connected to the N-terminal recombinant chitosanase mutant gene (without signal peptide sequence), and the Kex2 signal peptide cleavage site on the vector pPICZαA is used to cut off the secretory expression The chitosanase mutant signal peptide sequence was obtained to obtain a mature recombinant chitosanase mutant. The specific construction process of the pPICZαA-N-chitosanase mutant is as follows.
[0053]The purified PCR product was TA-ligated with the T vector pMD19-T, and the ligated product was transformed into E.coli Top10. Positive clones were obtained through blue-white screening and colony PCR identification. At the same time, the plasmid was extracted and sent to Beijing Liuhe Huada Gene Techno...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com