Controlling paddy rice cold-tolerant gene COLD2 and application thereof
A gene and rice technology, applied in the application field of rice cold tolerance breeding, can solve the problem of protein loss of function and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Embodiment 1, the discovery of cold tolerance of japonica rice Nipponbare:
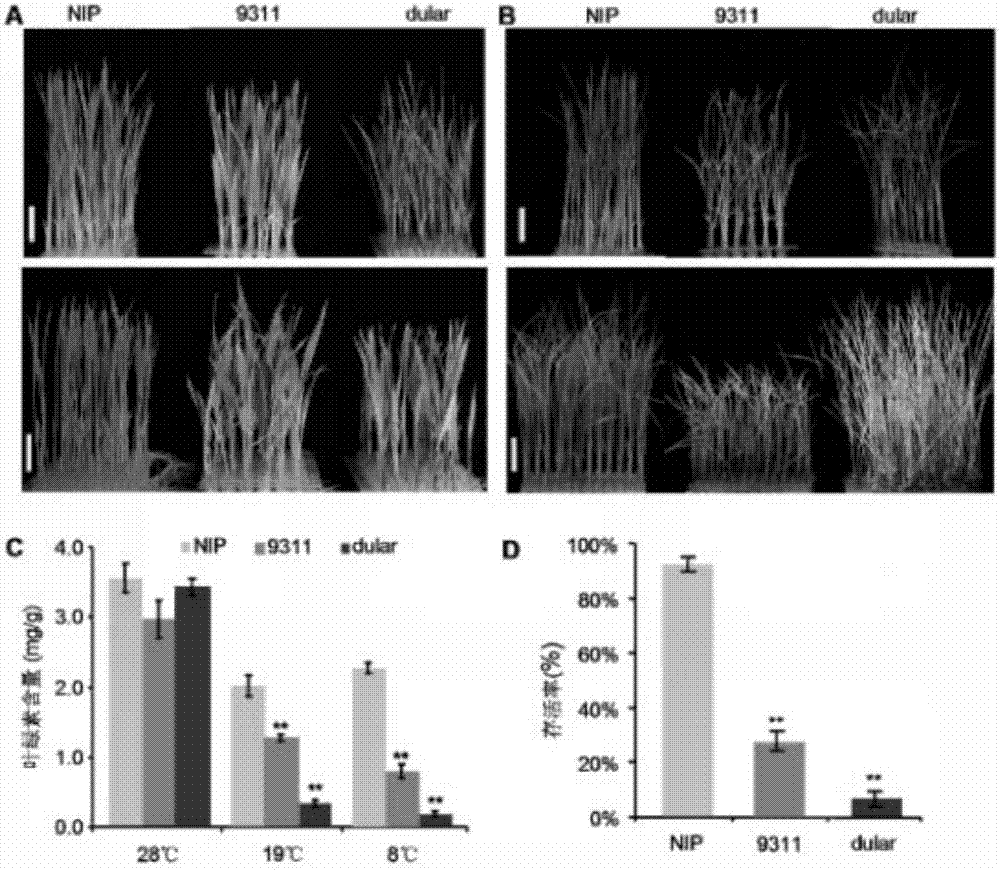
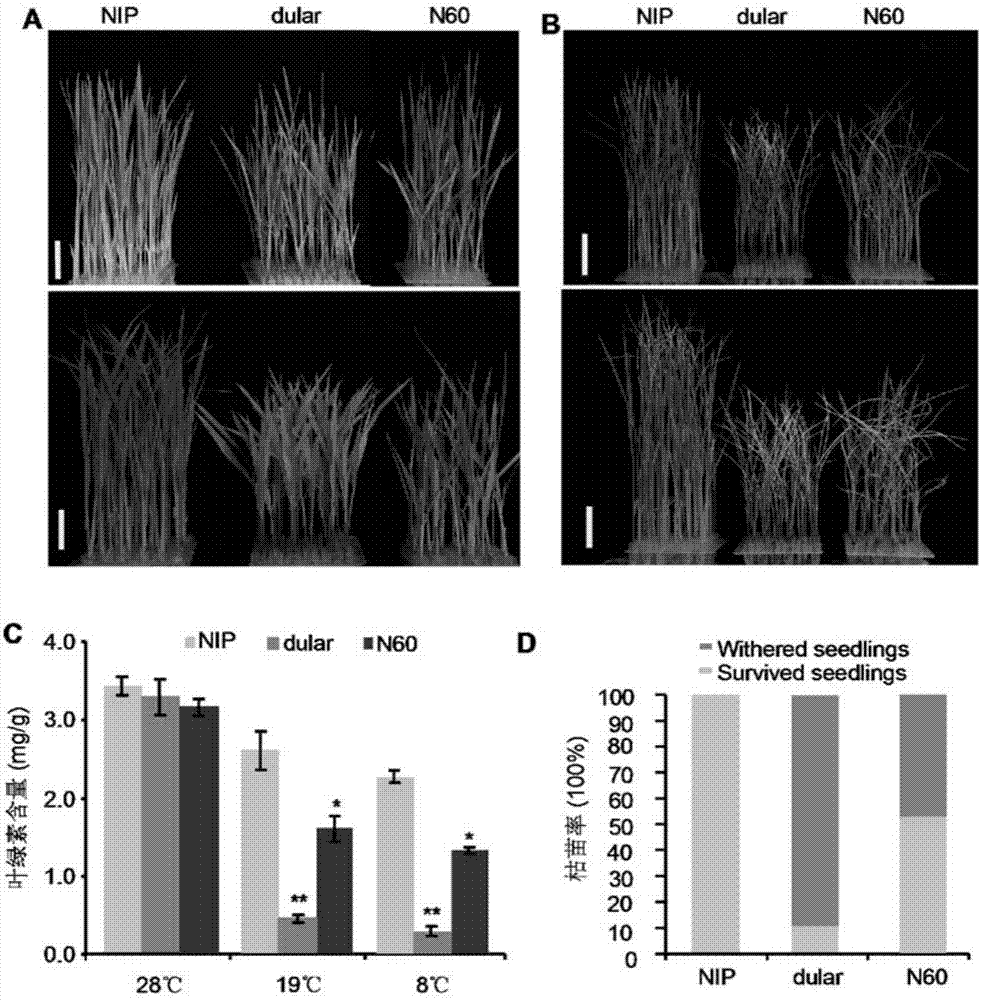
[0062] First, the cold tolerance of different rice varieties was analyzed. Among them, it was found that the low-temperature treatment of the japonica rice variety NIP and the indica rice variety 9311 and dual showed completely different cold tolerance. After the low-temperature treatment at 19°C, it was found that the leaves of NIP could still remain green, while 9311 and dual showed severe chlorosis and were not cold-tolerant phenotypes ( figure 1 A). Statistical analysis of differences before cold treatment found that there was no significant difference in chlorophyll content, and after cold treatment, the chlorophyll content in 9311 and dual leaves was significantly lower than that in NIP ( figure 1 C). In addition, after being treated at a lower temperature (8°C) for 7 days and then recovered at 28°C for 7 days, it was found that most of the NIPs were still alive, part of 9311 was alive,...
Embodiment 2
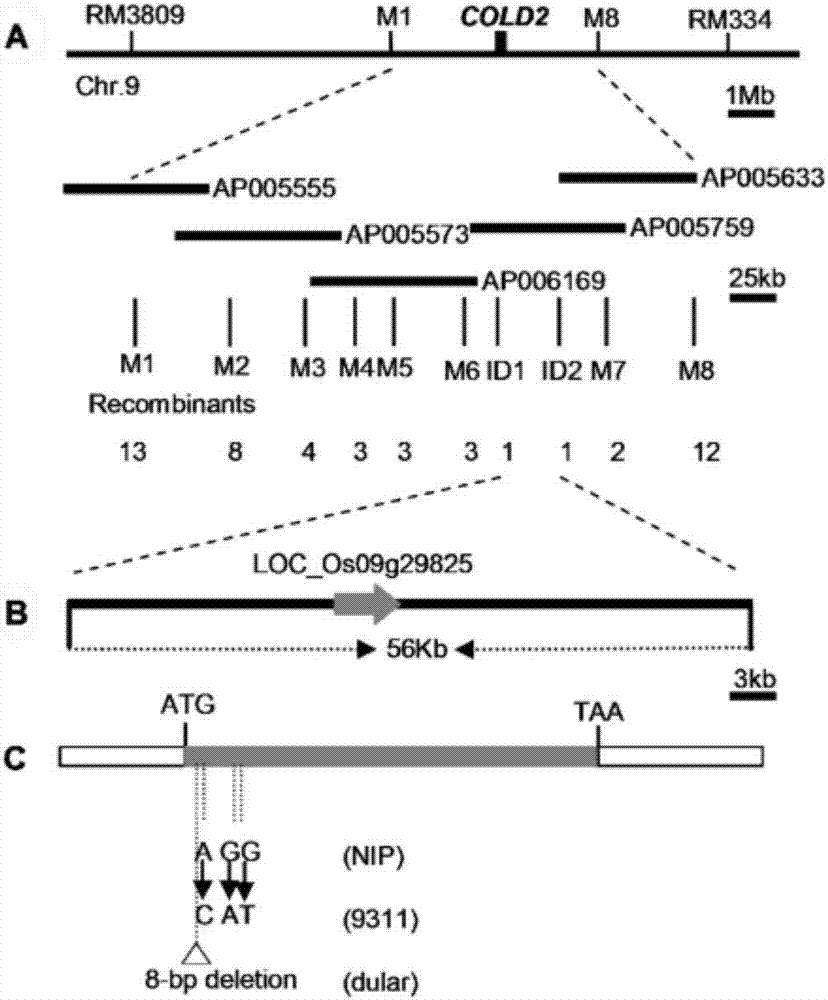
[0064] Embodiment 2, fine positioning of COLD2
[0065] The japonica rice Nipponbare NIP was crossed with the indica rice dual, F 1 All the plants exhibited cold tolerance, indicating that the COLD2 gene was controlled by a recessive nuclear gene. Will F 1 F 2 . Statistics F 2 Segregation population segregation ratio (Table 1), the results show that the segregation ratio of cold-tolerant plants and cold-sensitive plants is close to 3:1 segregation by chi-square test, which shows that COLD2 gene is controlled by a pair of single recessive nuclear genes.
[0066] Table 1. Genetic analysis of the cold tolerance gene COLD2
[0067]
[0068] Using 262 pairs of SSR primers evenly distributed in 12 chromosomes of rice preserved in our laboratory for polymorphism screening of NIP and dual, 180 pairs of SSR primers were screened for polymorphism. Then pick the F in 21 dual / NIP 2 Linkage analysis was performed on individual plants with cold-intolerant phenotype after low-tempe...
Embodiment 3
[0078] Embodiment 3, plant transformation
[0079] A total of 4.4-kb genomic DNA fragment from 5'-UTR 1880bp to 3'-UTR 831bp of COLD2 gene in Nipponbare was amplified. The primer sequences are:
[0080] F: GAATTC TGACATGTGGATCATGAACGTCACGAATCC
[0081] R: GGATCC CTTCATGTCTCATTCAACATCAACAACAGG
[0082]A 50 μL system was used for the PCR reaction system, and the PCR reaction system was configured according to the instructions of the high-fidelity polymerase KOD FX (Toyobo Company). The specific system was: 1 μL of DNA template, 25 μL of 2×PCR buffer, 1.5 μL, 10 μL of 2mM dNTPs, 1 μL of KOD FX enzyme, add ddH 2 Make up 50 μL of O;
[0083] The PCR amplification program is as follows: pre-denaturation at 94°C for 2 min; denaturation at 98°C for 10 s, annealing at 58°C for 10 s, extension at 68°C for 4 min, 35 cycles; final extension at 68°C for 10 min;
[0084] The obtained 4.4-kb genomic DNA fragment contains the sequence shown in SEQ ID NO:1.
[0085] Then it was conne...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com