Catechol 2,3-dioxygenase gene and application thereof
A catechol and dioxygenase technology, applied in the field of genetic engineering, can solve the problems of low enzyme activity and high temperature, and achieve the effect of solving environmental pollution problems and considerable economic benefits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Cloning of catechol 2,3-dioxygenase gene
[0044] (1) Extraction of total bacterial DNA
[0045] After the bacterial strain Sphingbium sp.MEA3-1 was cultured in LB liquid medium for 18 hours, the bacterial cells were collected by centrifugation at 5000r / min, and the total DNA of the bacterial cells was extracted using a genomic DNA extraction kit (Azanno Biotech), and the mass fraction was 0.75% agar Glycogel electrophoresis was used to detect the quality of the total DNA extracted.
[0046] (2) Primer design and synthesis
[0047] The upstream and downstream primers of catechol 2,3-dioxygenase were designed according to the genome information of Sphingbium bacteria, and the specific sequences are as follows:
[0048] Primer C23O-F: 5'- CATATG GCGATTCGTGGATTGCTC-3';
[0049] Primer C23O-R: 5'- CTCGAG GGTCAGAACGTTCAGAAAAGCT-3';
[0050] The underlined parts are NdeI and XhoI restriction sites respectively;
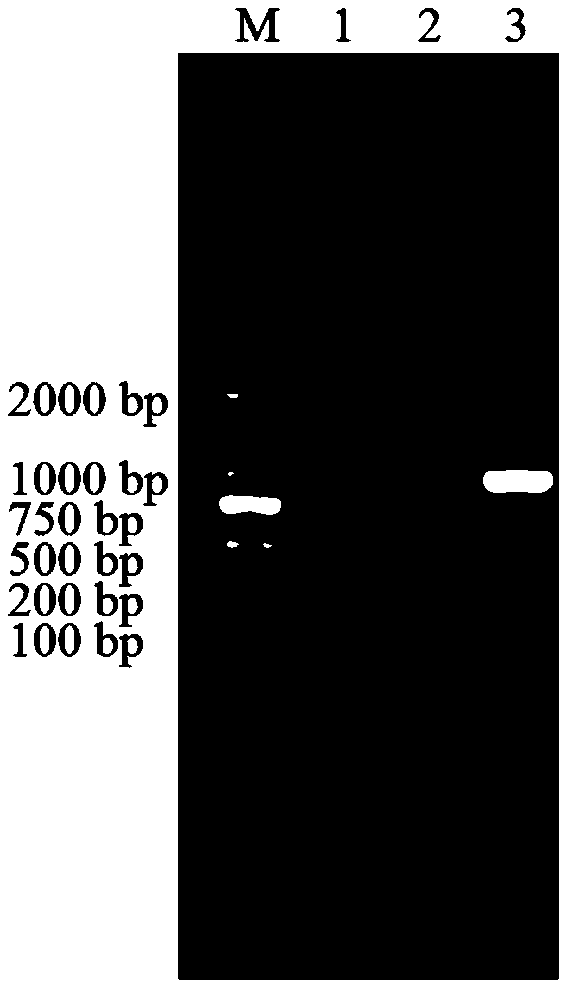
[0051] (3) PCR amplification of catechol dio...
Embodiment 2
[0064] Example 2 High-efficiency expression of recombinant catechol 2,3-dioxygenase protein
[0065] (1) Digest the positive clone plasmid successfully sequenced in Example 1 with NdeI and XhoI double enzyme digestion system (50 μL): NdeI 1 μL, Xho I 1 μL, 10×H buffer 5 μL, positive clone plasmid 20 μL, wxya 2 O 1 μL; react in a water bath at 37°C for more than 3 hours. After the digested product was detected by agarose gel electrophoresis with a mass fraction of 0.75%, a fragment of about 900 bp was recovered using a DNA gel recovery kit.
[0066](2) Referring to step (1), the vector pET-29a(+) (Merck-Novagen, Cat No. 69871) was double digested with NdeI and XhoI.
[0067] (3) Ligate the target fragment recovered in step (1) with the vector pET-29a(+) digested in step (2) to obtain the pET-29a(+) containing catechol 2,3-dioxygenase gene 29a(+) recombinant plasmid; the pET-29a(+) recombinant plasmid containing the catechol 2,3-dioxygenase gene linked by the enzyme was trans...
Embodiment 3
[0070] Example 3 Purification and concentration of recombinant catechol 2,3-dioxygenase
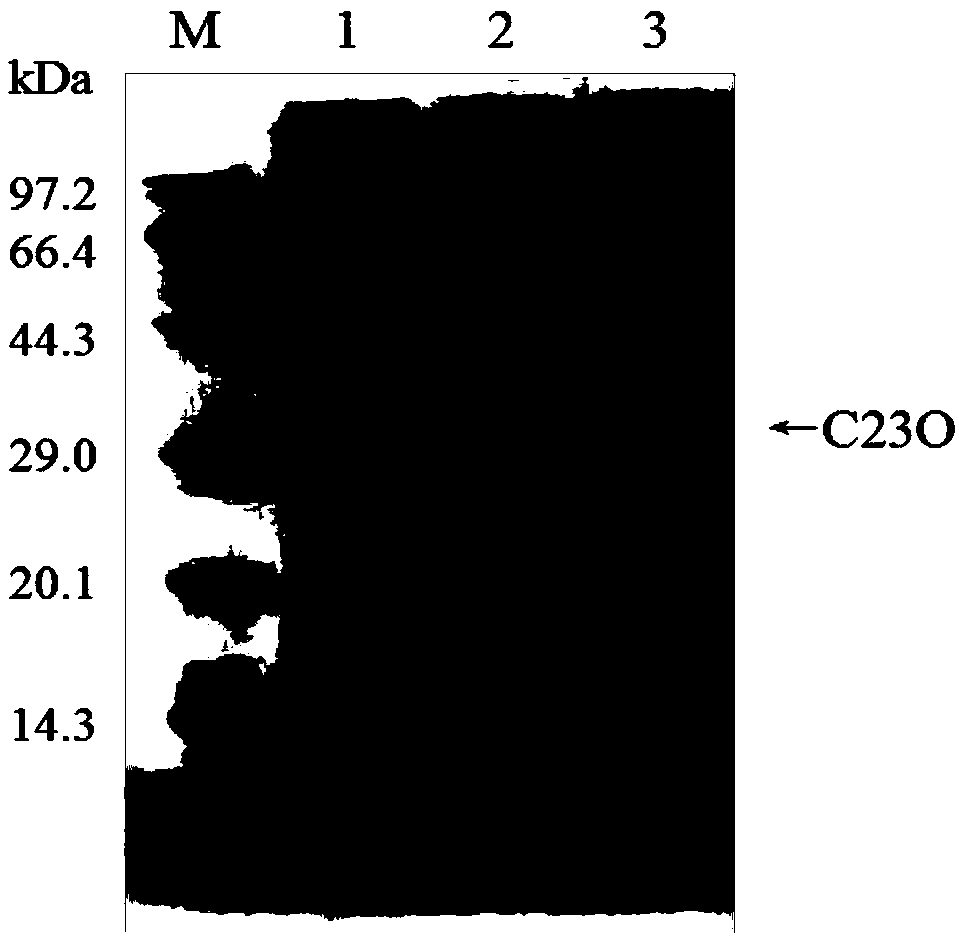
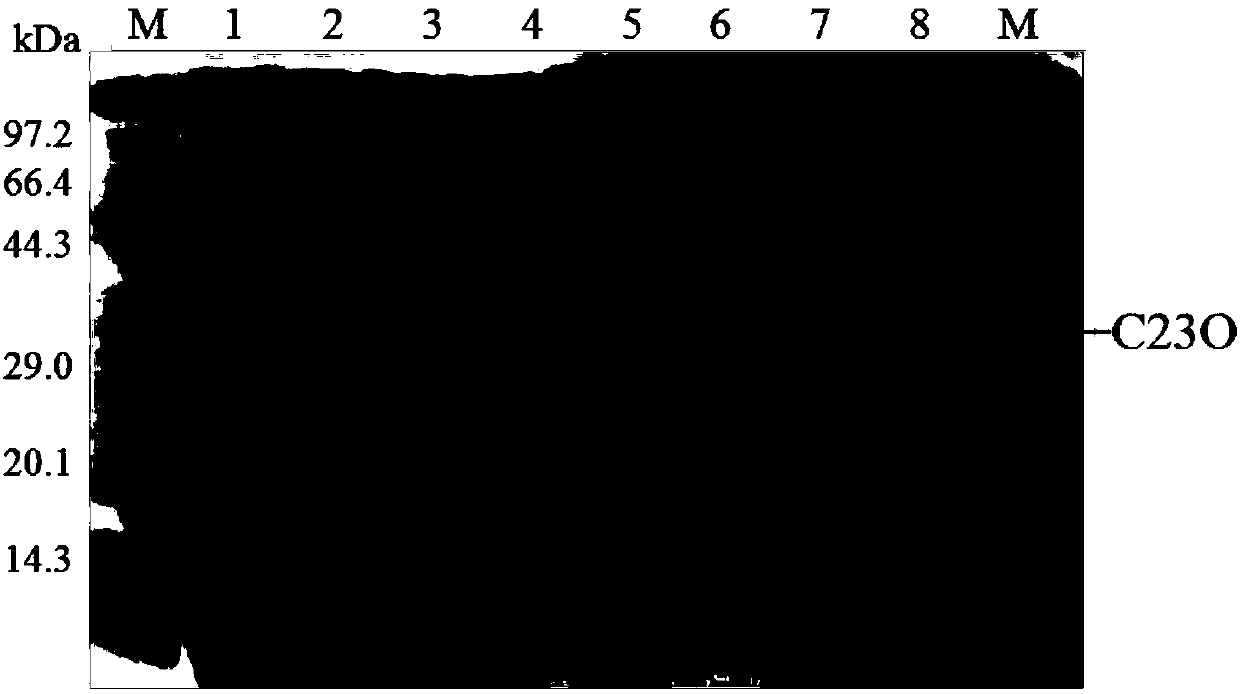
[0071] Using the six histidine tags introduced during primer design, the crude enzyme solution (prepared in step (4) of Example 2) that induces expression of catechol 2,3-dioxygenase was passed through Ni-NTAResin (Sangon Biotech) Purify the affinity chromatography column, the steps are as follows: first equilibrate the column with 10mL buffer solution A (20mM PBS pH 8.0, 300mM NaCl, 0mM imidazole), take 2.5mL crude enzyme solution and load it on the Ni-NTA affinity chromatography column, Elute with 5mL buffer solution A, and then use 5mL buffer B (20mM PBS pH 8.0, 300mM NaCl, 20mM imidazole), buffer C (20mM PBS pH 8.0, 300mM NaCl, 50mM imidazole), buffer D (20mM PBS Gradient elution was performed with buffer E (20 mM PBS pH 8.0, 300 mM NaCl, 100 mM imidazole), buffer E (20 mM PBS pH 8.0, 300 mM NaCl, 250 mM imidazole) and buffer F (20 mM PBS pH 8.0, 300 mM NaCl, 500 mM imidazole). Use S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com