Tumor acidity responsive autophagy-induced polypeptide as well as preparation method and application thereof
An acidity-responsive and autophagy technology, applied in biochemical equipment and methods, anti-tumor drugs, chemical instruments and methods, etc., can solve the problems of limited application, lack of tissue specificity, poor in vivo stability, etc. High rate and strong antitumor activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: Construction of Fusion Protein Particles
[0022] Chemical synthesis of pHLIP-Beclin 1 gene sequence
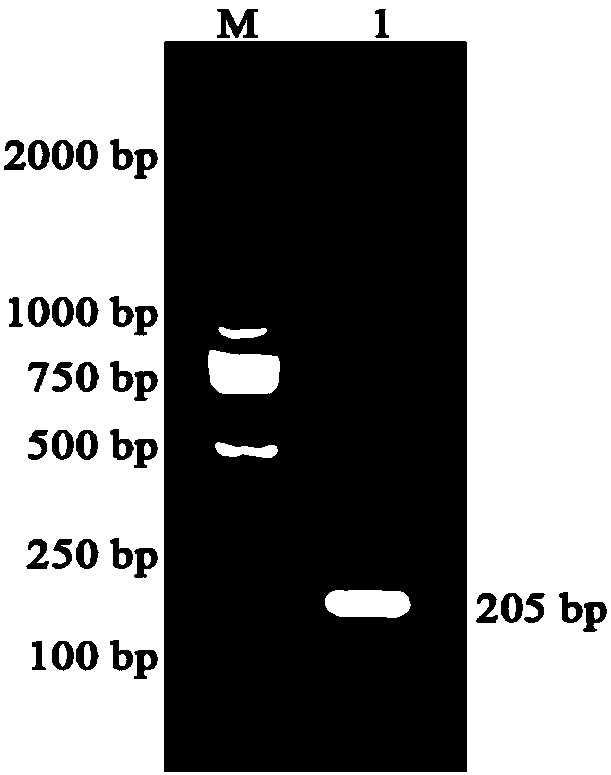
[0023] GGATCCGCGGCGGAACAGAACCCGATTTATTGGGCGCGCTATGCGGATTGGCTGTTTACCACCCCGCTGCTGCTGCTGGATCTGGCGCTGCTGGTGGATGCGGATGAAGGCACCTGCGGCACCAACGTGTTTAACGCGACCTTTCATATTTGGCATAGCGGCCAGTTTGGCACCTAACT CGAG, and at the two ends of the target protein gene sequence were designed BamH I and Xho enzyme cutting sites. Then, the target protein gene is connected into the pET-32a vector through the above-mentioned restriction site, so as to obtain the target protein expression vector. (See figure 1 and 2)
Embodiment 2
[0024] Embodiment 2: Expression and purification of fusion protein
[0025] (1) Expression of fusion protein
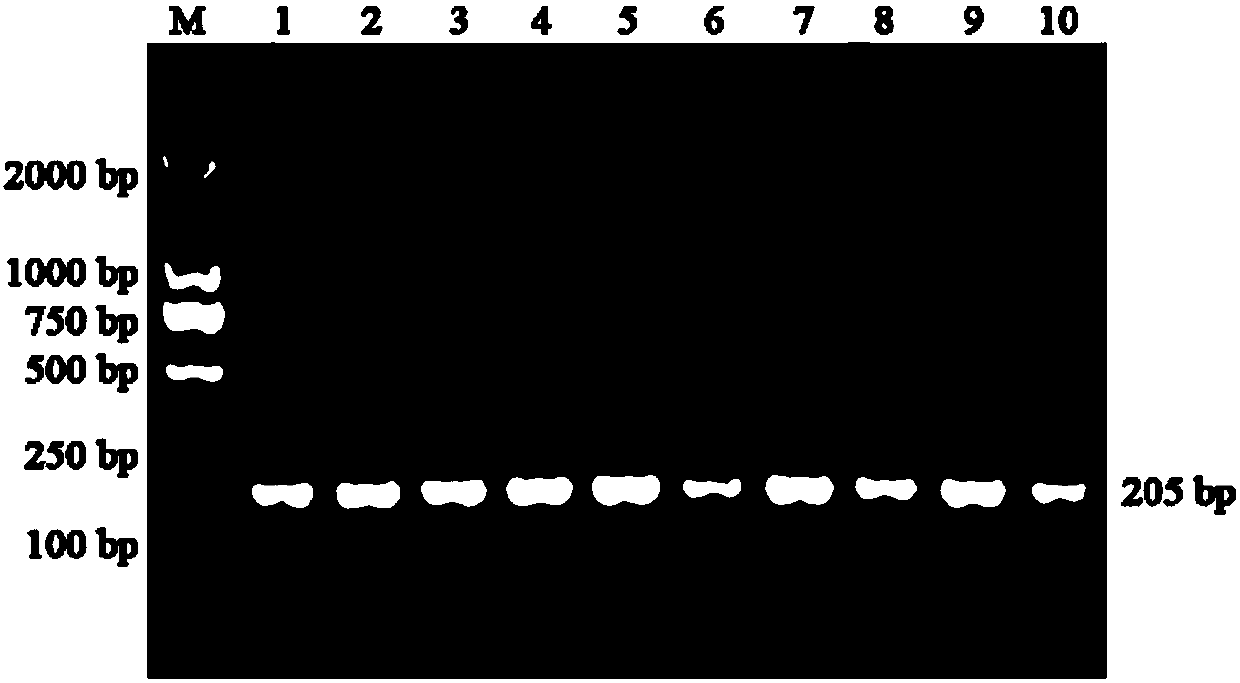
[0026] The above fusion protein expression vector was transformed into BL21(DE3) Escherichia coli, and 50 μL of bacterial liquid was added to 5 mL of LB liquid medium, and cultured on a shaking table at 37° C. at 200 rpm for 8 hours. The bacterial liquid was transferred to 500 mL LB liquid medium, cultivated to OD=0.6-0.8 at 37° C., 200 rpm, and induced to express overnight with IPTG (1 mM) at 16° C., 200 rpm. (See image 3 )
[0027] (2) Purification of fusion protein
[0028] Centrifuge (8000rpm, 10min) the above-mentioned IPTG-induced bacterial solution, discard the supernatant, and harvest the bacteria. The precipitate was blown away with 10 mM PBS (pH=7.4) solution, sonicated, centrifuged at 13000 rpm for 20 min, and the supernatant was collected. The supernatant was purified by Ni ion affinity chromatography. After the total protein was fully adsorbed, the ...
Embodiment 3
[0029] Example 3: Crystal violet staining method to detect the effect of autophagy-inducing polypeptide on cell proliferation
[0030] MCF-7 cells in the exponential growth phase were treated with 5×10 3 per well into a 96-well culture plate at 37°C with 5% CO 2 After incubation in the incubator for 24 hours, different concentrations of tagged protein Trx, Beclin 1, and fusion protein were added to the fresh culture medium at pH=7.4 / 6.5 to the final concentrations of 0 μM, 10 μM, 50 μM, and 100 μM, and five replicates were set for each concentration. hole. After incubation for 24 hours, the liquid in the wells was aspirated, washed twice with PBS buffer, and 50 μL of 0.5% crystal violet solution was added to each well. After staining at room temperature for 20 min, the liquid in the wells was aspirated, lightly washed twice with PBS buffer, and washed with BioTek Cytation 5. Hole imager, observed under 4x microscope. After observation, 200 μL of methanol solution was added to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com