DNA (Deoxyribonucleic Acid) methylation detection kit and application method thereof
A detection kit and methylation technology, applied in the field of biological analysis, can solve the problems of time-consuming and low sensitivity, and achieve the effect of simple operation and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1 A kind of DNA methylation detection kit
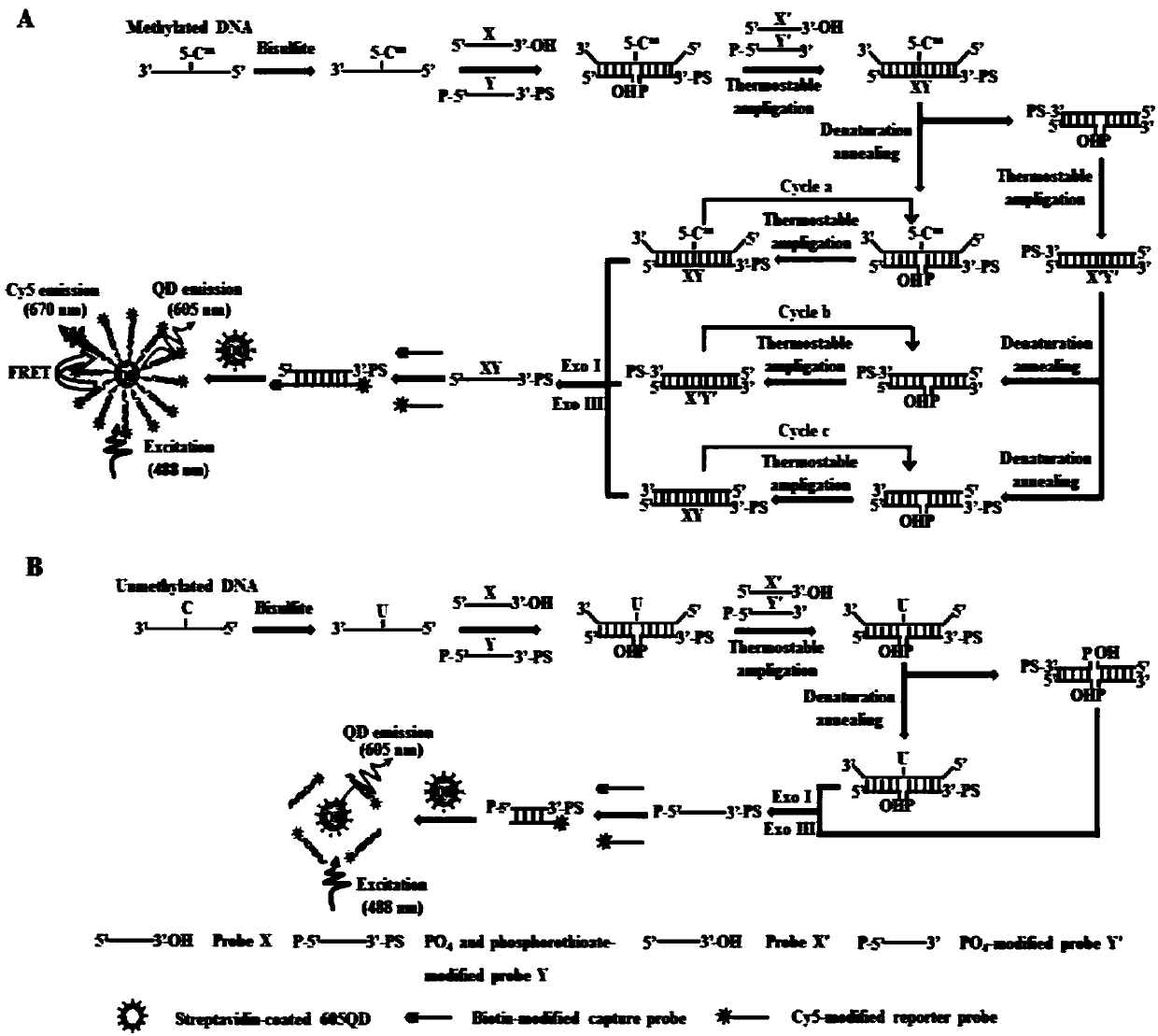
[0048] The kit includes: sodium bisulfite, hydroquinone, desalting column, 0.3 mole per liter of sodium hydroxide, ammonium acetate, ethanol, trisaminomethane hydrochloride, potassium chloride, magnesium chloride, nicotinamide adenine Dinucleotide, polyethylene glycol octylphenyl ether, probe X, probe Y, probe X', probe Y', exonuclease I, exonuclease III, 10×NEBufferI, sulfuric acid Ammonium, Cy5-labeled reporter probe, biotin-labeled capture probe, thermostable ligase, and 605 quantum dots.
[0049] The nucleotide sequences of the probe X, probe Y, probe X' and probe Y' are shown in SEQ ID NO.1-4; the probe Y is modified with a phosphate group at the 5' end, and the 3' The end is modified by phosphorothioation, and the probe Y' is modified with a phosphate group at the 5' end.
[0050] The Cy5-labeled reporter probe is: 5'-TGA CTC TGT GGA GTC CTG CC-3', as shown in SEQ ID NO.5, wherein the fourth base T base at t...
Embodiment 2
[0051] The using method of DNA methylation detection kit described in embodiment 2
[0052] The method for using the DNA methylation detection kit is divided into the following steps:
[0053] (1) DNA bisulfite treatment, after denaturing the DNA to be tested at 42°C and 0.35 moles per liter of sodium hydroxide for 30 minutes, carry out sodium bisulfite reaction: add the denatured DNA to be tested into fresh The prepared 3.2 moles per liter of sodium bisulfite and 0.5 millimoles per liter of hydroquinone were reacted at 50°C for 16 to 18 hours; after the reaction, DNA was recovered using a desalting column, and 0.3 moles per liter of sodium hydroxide was used to React at 37°C for 15 minutes to complete the modification, then neutralize with ammonium acetate, ethanol precipitate and dry, and store the DNA solution in ultrapure water at 20°C for later use;
[0054] (2) Three-step circular ligase chain reaction, the reaction system is: 20 millimoles per liter of trisaminomethane...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com