Novel pullulanase and coding gene and application thereof
A new pullulanase and gene technology, applied in the field of new pullulanase and its coding gene and its application, can solve the problems of few microorganisms and difficult microbial sampling
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Cloning of Neopulluanase Neopullu31369 gene and construction of expression vector.
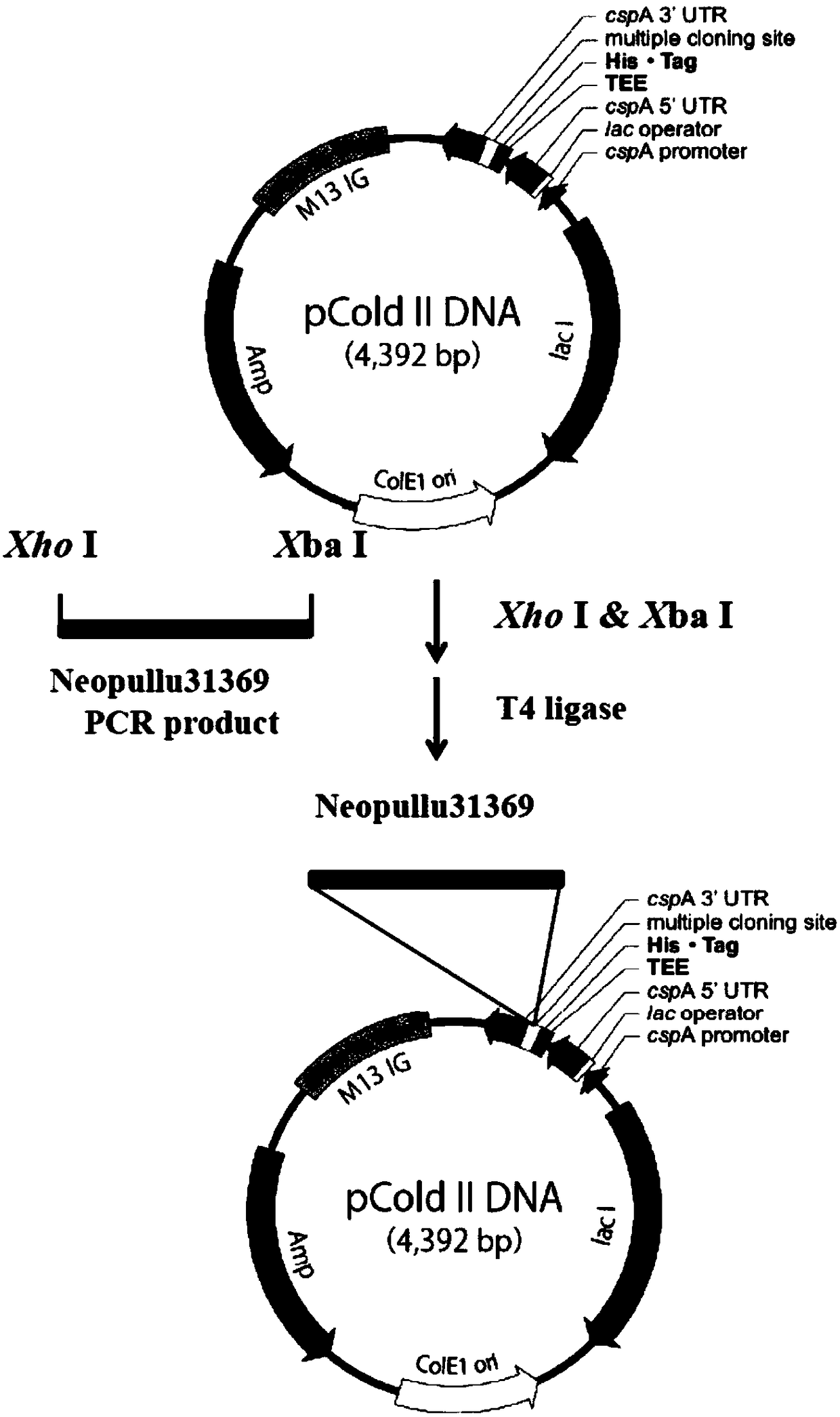
[0034] First, annotate and analyze the sequencing results of the metagenomic library of the deep-sea sediment sample E406 (water depth 3318, longitude 119°44.966'E, 18°44.922'N) to obtain the gene information of the new pullulanase; Gene sequence information, design specific primers, use the metagenomic library as a template, carry out PCR amplification, and obtain the Neopullu31369 coding gene fragment, the full length is 1767bp, which is consistent with the expected size (see figure 1 ). Such as figure 2 As shown, the recovered and purified Neopullu31369 gene PCR product was cut with XhoI and XbaI, and then ligated with the linearized vector pCold cut with XhoI and XbaI to form the expression vector pCold-Neopullu31369 containing the Neopullu31369 gene fragment.
[0035] The purified pCold-Neopullu31369 recombinant plasmid was sequenced bidirectionally with T7 and T7t pr...
Embodiment 2
[0050] Example 2: Recombinant expression and purification of neopullulanase Neopullu31369.
[0051] The recombinant expression plasmid with correct sequencing was transformed into the expression host strain BL21(DE3) to construct an engineering strain. Pick a single colony and inoculate it in Amp + In the LB liquid medium, 37 ℃ shaking culture overnight, as the seed fungus. First use 50% glycerol to preserve the seed, then take the remaining bacterial solution and inoculate it in fresh Amp at a volume ratio of 1:100. + In LB medium, shake vigorously at 37°C to amplify the culture until OD600 is about 0.6-0.8, add IPTG to a final concentration of 0.1mmol / L, and induce expression overnight at 18°C. Centrifuge at 4°C and 7500rpm for 10 minutes to collect the bacteria, and reserve 1ml of the bacteria liquid for SDS-PAGE analysis. After washing the cells with pre-cooled TE buffer (pH8.0), resuspend the cells with pre-cooled ultrasonic buffer. With 25% power, sonicate in an ice ...
Embodiment 3
[0052] Example 3: Determination of the enzyme activity of the new pullulanase Neopullu31369 on different substrates.
[0053] (1) DNS method
[0054] Take 2 test tubes, namely the control tube and the sample tube. Add 800 μl of Tris buffer solution and 100 μl of substrate solution to each of the two test tubes, incubate in a 37°C water bath for 5 minutes, then add 100 μl of inactivated purified enzyme solution (boiling in a boiling water bath for 10 minutes) to the control tube, and add 100 μl of purified enzyme solution to the sample tube , mixed immediately and timed, reacted accurately in a 37°C water bath for 30 minutes, then added an equal volume of DNS to terminate the reaction, developed color in a boiling water bath for 5 minutes, and measured the absorbance value with a 540nm spectrophotometer after cooling, repeated three times for each sample, and took the average value.
[0055] Enzyme activity unit (1U) is defined as the amount of enzyme required to catalyze the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com