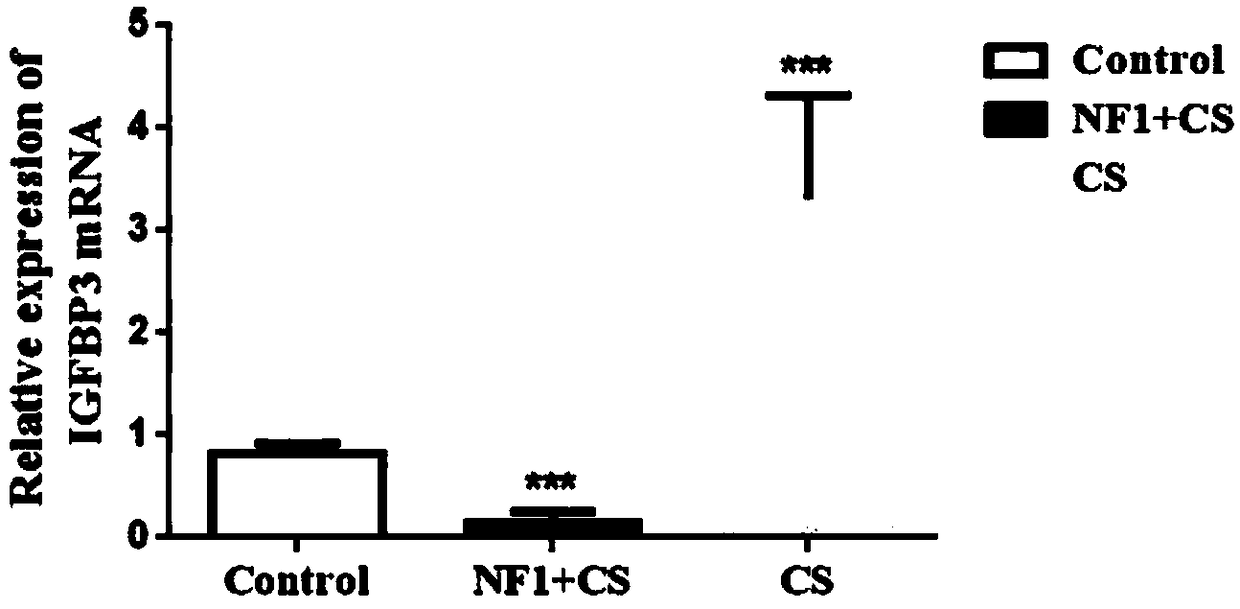
Application of IGFBP3 (insulin-like growth factor-binding protein 3) in preparation of products to diagnose type I neurofibromatosis with spinal deformity
A technique for neurofibromas, spinal deformities, used in biotechnology and medical testing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] The collection of embodiment 1 sample and the arrangement of sample data
[0031] From January 2017 to December 2017, peripheral blood samples from 5 patients with neurofibroma combined with spinal deformity and 5 relatively normal peripheral blood samples were obtained. All patients did not receive chemotherapy or radiotherapy before the operation, and the informed consent of the patients was obtained before the operation, which was reviewed by the ethics committee.
[0032] Sample processing: EDTA anticoagulated whole blood was mixed with Trizol at a ratio of 1:1, mixed thoroughly and placed in a 1.8ml cell cryopreservation tube, cooled rapidly in liquid nitrogen for 30 seconds and stored in a -80°C refrigerator.
Embodiment 2
[0033] Example 2 Peripheral blood RNA extraction
[0034] 1. Homogenization treatment
[0035] Take fresh blood, add 3 times the volume of erythrocyte lysate, mix well, place at room temperature for 10 minutes, and centrifuge at 10,000 rpm for 1 minute. Discard the supernatant and collect the white blood cell pellet. Add 1ml Trizol to every 100-200ul blood collected leukocyte pellet.
[0036] 2. Layering
[0037] After the sample was added to Trizol, it was left at room temperature for 5 minutes to fully lyse the sample.
[0038] Add 200ul of chloroform to every 1ml of Trizol, vortex vigorously and mix well, then place at room temperature for 3-5min to allow natural phase separation.
[0039] 3. RNA precipitation
[0040] Centrifuge at 12000rpm at 4°C for 10-15min. The sample will be divided into 3 layers: yellow organic phase, intermediate layer and colorless aqueous phase. RNA is mainly in the aqueous phase. Transfer the aqueous phase (usually absorbing 550-600ul) to a n...
Embodiment 3
[0051] Example 3 RNA-seq sequencing library construction and quality inspection of peripheral blood RNA
[0052] 1. Data quality assessment
[0053] The construction and sequencing of the cDNA library was entrusted to Beijing Novogene Technology Co., Ltd. to complete. The sequencing data of 10 samples were checked for sequencing error rate, GC content distribution and raw data filtering to obtain clean reads for subsequent analysis. The data summary is shown in Table 2.1. It is generally considered that RNA-seq data is used for gene differential expression analysis between different libraries, the total number of reads in the library is at least 10M, the overall GC content of the data is kept at 40%-60%, and the Q30 is more than 80%. In the data obtained by this sequencing, clean bases accounted for more than 7.2G, Q2 bases accounted for more than 95.18%, Q3 bases accounted for more than 89.15%, and the GC content remained stable between samples, ranging from 54.29% to 56.80%...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com