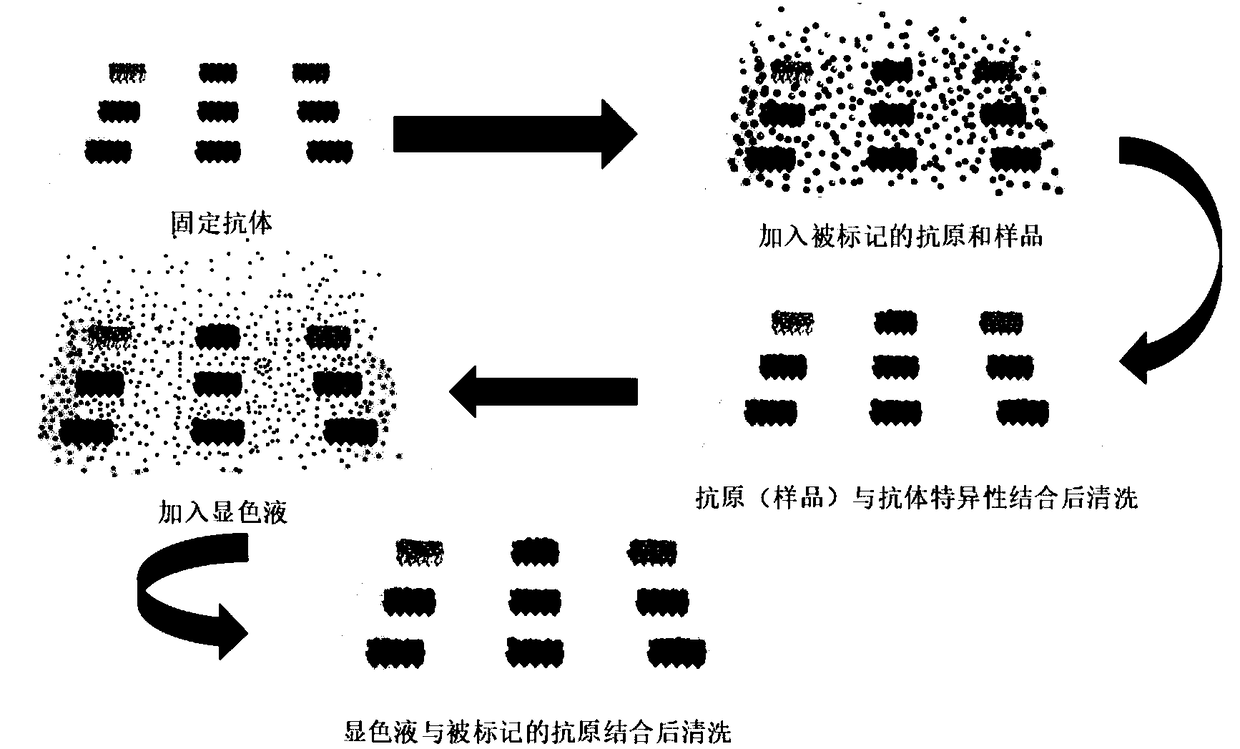
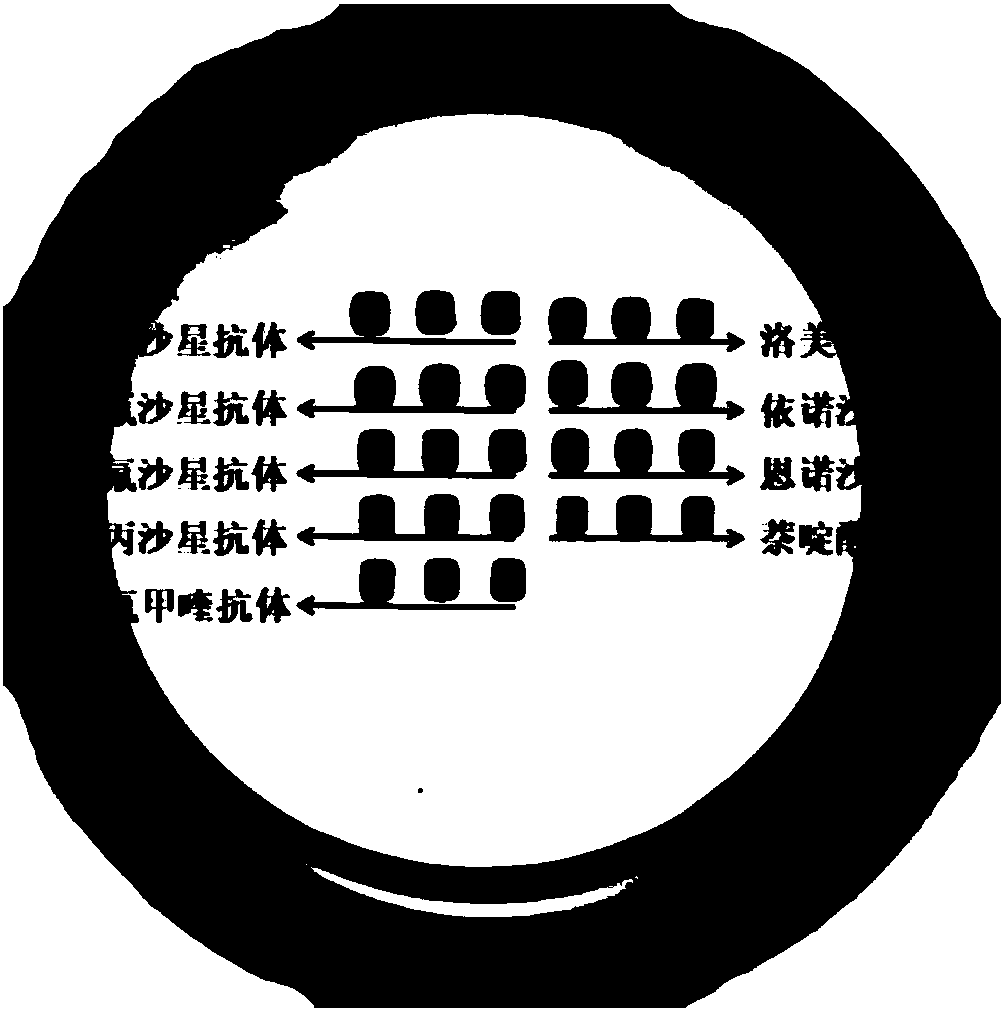
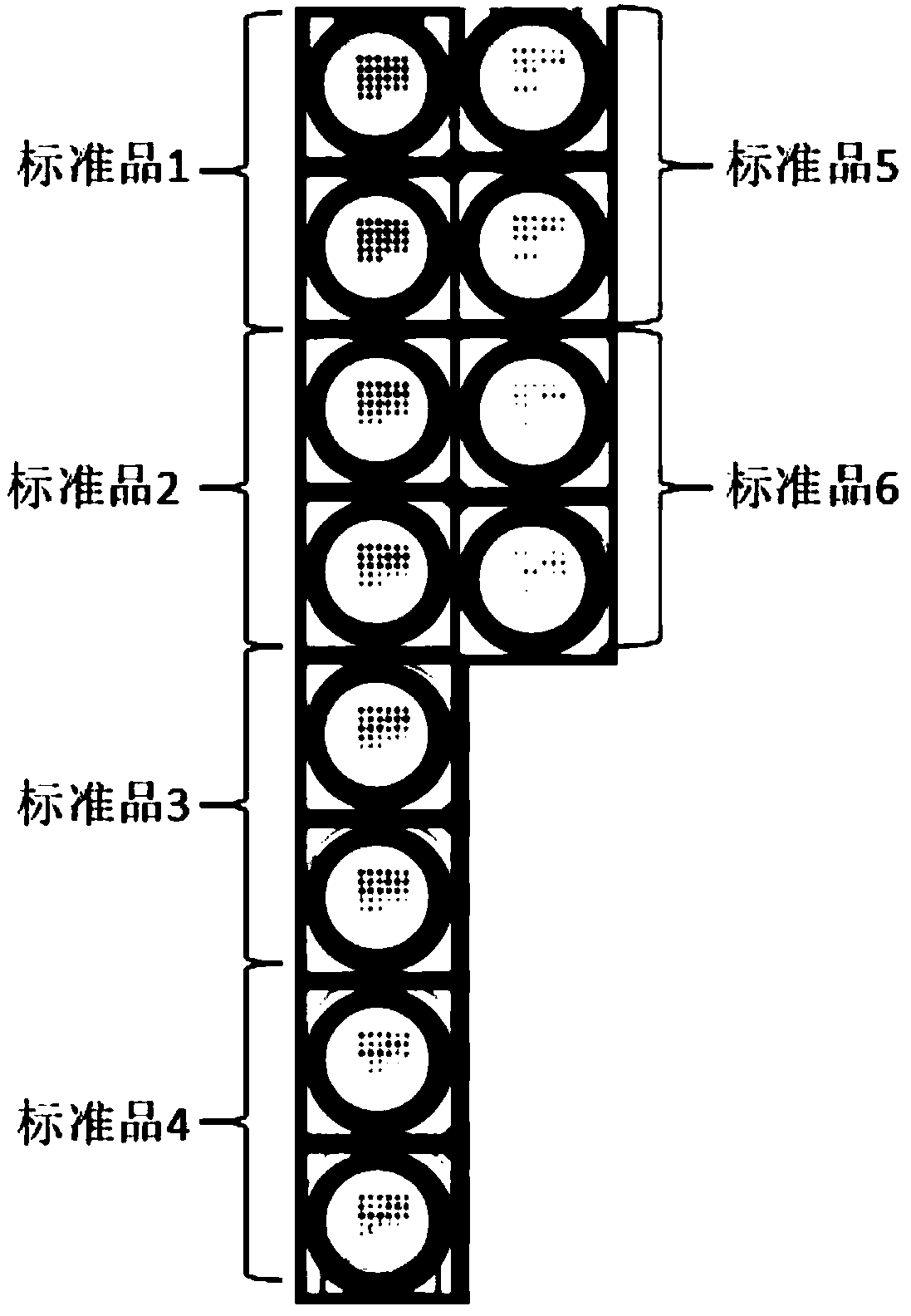
Method for simultaneously detecting residual amount of quinolone antibiotics
A technology for quinolones and antibiotics, applied in the field of simultaneous detection of quinolone antibiotics residues, can solve the problems of complex technical operations, expensive equipment, high detection limits, etc., and achieve the effects of improving detection efficiency, shortening detection time, and simple experimental operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] A method for simultaneously detecting 9 kinds of quinolone antibiotic residues:
[0037] (1) Preparation of visualized protein chip: prepare a 6×5 matrix on a 96-well plate with a biochip spotter, and spot nine quinolones (ofloxacin, lomefloxacin, pefloxacin, Enoxacin, norfloxacin, enrofloxacin, ciprofloxacin, nalidixic acid, flumequine) monoclonal antibody, the sample volume is 50nL / point, and each sample is repeated 3 points; After the end, incubate in a 37°C incubator for 2 hours to immobilize the antibody on the bottom of the plate; add 200 μL chip blocking agent (BSA) to each well, incubate in a 37°C incubator for 2 hours, and then wash with washing buffer 3 times. Each time for 10 seconds, pat dry, and store in a 4°C refrigerator for later use;
[0038] (2) Add 25 μL (ofloxacin, lomefloxacin, pefloxacin, enoxacin, norfloxacin, enrofloxacin, cyclofloxacin, Profloxacin, nalidixic acid, flumequine) mixed standards or samples with different concentration gradients, ...
Embodiment 2
[0049] Specificity test for quinolones
[0050] The specificity of quinolone antibodies is usually judged using competition inhibition curves. Different concentrations of antigen and interferent were used to calculate their respective binding ratios (B / B 0 ), draw competitive inhibition curves, and calculate their respective ICs 50 . The cross-reactivity rate was calculated according to the following formula (1).
[0051] CR=[IC 50 (analyte) / IC 50 (interference object)]×100% (1)
[0052] IC 50 is the concentration required for 50% inhibition of the analyte or interferent-induced signal.
[0053] The experimental results are shown in Table 2, indicating that the cross-reactivity rates between quinolone antibodies and nine quinolone drugs are all between 1% and 5%. It shows that the specificity of the quinolone antibody is good, there is no crossover, and each quinolone antibiotic can be detected separately.
[0054] Table 2 Quinolones cross-reaction rate
[0055]
...
Embodiment 3
[0058] Establishment of standard curve for quinolones
[0059] The indirect competition method was used to explore the standard curve of quinolones. 3 parallel microarray points for each antibody concentration point, the signal value is taken as the average value, within a certain range, with the increase of the concentration of the competition standard, the content of the labeled artificial antigen bound by the fixed antibody will be less and less, Therefore, the detected signal value will be lower. After exceeding this range, the detected signal value will not change with the increase of the concentration of the competition standard. Use standard products of ofloxacin, standard products of lomefloxacin, standard products of pefloxacin, standard products of enoxacin, standard products of norfloxacin, standard products of enrofloxacin, standard products of ciprofloxacin, naphthalene Glycolic acid standard substance, flumequine standard substance (as shown in table 3) mixed st...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com