Primer and probe used for detecting EGFR TK1 sensitive mutant
A probe, sensitive technology, applied in the fields of biotechnology and medicine, can solve the problems of increased detection cost, stay, high complexity, etc., to achieve the effect of reducing detection sites, reducing detection cost, and reducing total time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Detection limit, sensitivity and specificity of detection primers and probes
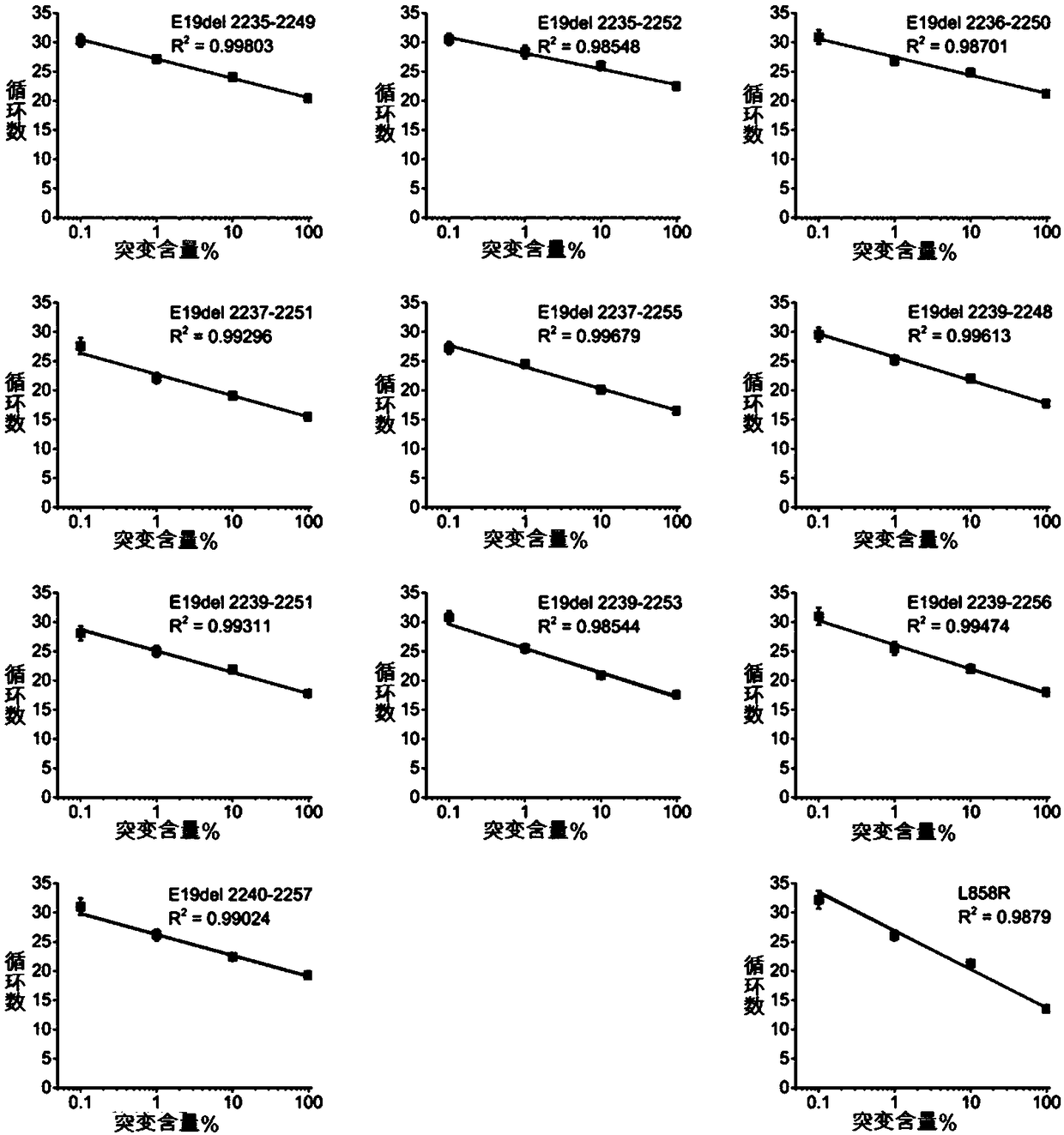
[0036] Construct plasmids containing 19 exon deletion mutations and L858R mutations respectively. The corresponding construction sequences in the plasmids are shown in Table 7, including 10 exon 19 exact mutations, L858R mutations, 19 exon wild type, and L858R wild type . Use the L858R wild-type plasmid sample with 19 exons to dilute the mutant plasmid by 1 fold, 10 fold, 100 fold, and 1000 fold, and then use the primers, probes and PCR kits designed above for detection to obtain each detection The lowest detection limit of the site. The result is figure 1 As shown, the minimum detection limit for each mutation site is 1%. When the number of cycles is 27-31, as shown in the figure, the mutation rate is between 0.1% and 1%. At this time, the mutation is suspected to be positive, and DNA needs to be extracted again for repeated testing.
[0037] Table 7 Plasmid detection sequence
[0038]
[0039]...
Embodiment 2
[0044] Detection of EGFR TKI sensitivity mutations in blood cfDNA of lung cancer patients. The experimental method is as follows:
[0045] 1. Take 5ml of lung cancer patient's blood. Centrifuge at 2500 rpm for 10 minutes and take 1 ml of upper plasma.
[0046] 2. Add 200μl of 5% SDS lysate to plasma.
[0047] 3. Add an equal volume of saturated phenol solution, gently shake for 5 minutes, centrifuge at 5000g for 15 minutes, and take the supernatant.
[0048] 4. Add an equal volume of chloroform to the supernatant, shake for 5 minutes, centrifuge at 3000g for 10 minutes, and take the supernatant.
[0049] 5. Add three times the volume of frozen ethanol to the supernatant and mix, centrifuge at 12,000 rpm for 2 minutes, and the DNA will sink to the bottom of the tube. Carefully discard the liquid.
[0050] 6. Drain the remaining liquid at the bottom of the tube and add 50μl TE to dissolve the DNA pellet.
[0051] 7. Use the above primers, probes and PCR kit to detect EGFR TKI sensitive mu...
Embodiment 3
[0054] Detection of EGFR TKI sensitivity mutations in tissue DNA of lung cancer patients. The experimental method is as follows:
[0055] 1. Take fresh tissue the size of soybeans, add 200μl of 10mg / ml proteinase K solution, and incubate at 70°C for 1h.
[0056] 3. Add an equal volume of saturated phenol solution, gently shake for 5 minutes, centrifuge at 5000g for 15 minutes, and take the supernatant.
[0057] 4. Add an equal volume of chloroform to the supernatant, shake for 5 minutes, centrifuge at 3000g for 10 minutes, and take the supernatant.
[0058] 5. Add three times the volume of frozen ethanol to the supernatant and mix.
[0059] 6. Add the mixed liquid to the DNA separation column, centrifuge at 12000 rpm for 1 min, and discard the lower liquid.
[0060] 7. Add 500 μl of 75% ethanol solution to the separation column, centrifuge at 12000 rpm for 1 min, and discard the lower liquid.
[0061] 8. Place the spin column in the open to evaporate the ethanol, add 50μl TE to elute the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com