Set of genes used for bladder cancer detection and application thereof
A technique for bladder cancer, genetics, used in cancer detection and molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0114] Collection and processing of training set samples;
[0115] The present invention analyzed 52 cases of bladder cancer patients, 40 cases of non-bladder cancer patients with other urinary system diseases, a total of 92 cases of clinical data of urine samples and 20,500 gene expression abundance data, and constructed bladder cancer genes in urine express database.
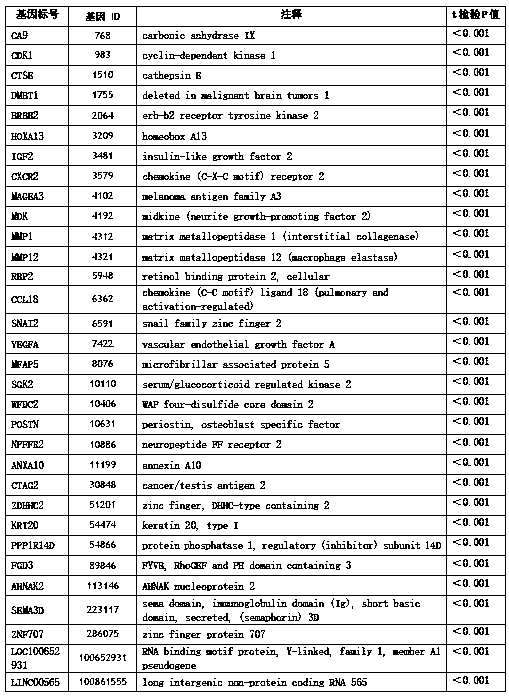
[0116] Screening of 32 specific genes;
[0117] According to the measured value of gene expression abundance, the inventors selected 32 genes closely related to bladder cancer from 20,500 genes by using the statistical analysis method T-test. These genes were differentially expressed in bladder cancer, which was statistically significant, as shown in Table 1.
[0118] Table 1: 32-gene set
[0119]
[0120] Construction of 32-gene statistical analysis model:
[0121] Based on the expression patterns of 32 specific genes in bladder cancer and non-bladder cancer samples, the inventors used the Support Vector...
Embodiment 2
[0123] In this example, the inventors analyzed the high-throughput sequencing data of 107 urine samples, including 63 bladder cancer samples and 44 non-bladder cancer samples. Each sample was discriminated by the 32-gene statistical analysis model, and compared with the clinical diagnosis results, the accuracy rate was 91.6%, the sensitivity was 92.1%, and the specificity was 90.9%, see Table 2.
[0124] Table 2: Discrimination results of 32-gene detection in the validation set of 107 cases
[0125]
Embodiment 3
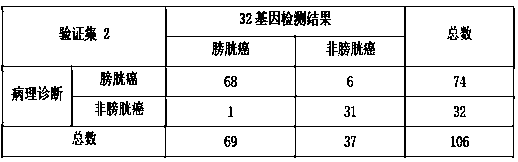
[0127] In this example, the inventors collected 114 urine samples, including 80 bladder cancer samples and 34 non-bladder cancer samples. The researchers first centrifuged the exfoliated cells in the urine, and then used the purification column extraction method instead of the traditional TRIzol method to extract total RNA. Among them, 8 samples were excluded due to unqualified RNA quality, and the rejection rate was 7%. Ultimately, the study included 106 samples, including 74 bladder cancer samples and 32 non-bladder cancer samples. The extracted and purified RNA was reverse-transcribed to obtain cDNA, and the real-time quantitative polymerase chain reaction of 32 genes was performed to detect the gene expression level in the exfoliated cells in urine, and the analysis model was used to calculate the similarity score between the sample and bladder cancer.
[0128] International patent application WO2014 / 118334 describes the use of TRIzol (Invitrogen, Carlsbad, CA, USA) extra...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com