Nucleic acids and method for synchronous detection and distinguishing of five canine diarrhea viruses and kit
A diarrhea virus, simultaneous detection technology, applied in biochemical equipment and methods, microbial determination/inspection, resistance to vector-borne diseases, etc. Viruses, multiple methods detection effect is not ideal and other problems, to achieve the effect of fast reaction, simple operation and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Design and synthesis of embodiment 1 primers and probes
[0041] The genome sequences of standard strains of CAV1, CAV2, CCV, CDV, and CPV were downloaded from the NCBI website, and the conserved regions of the standard strains of each virus were selected by DNAStar software, and primers were designed by Primer5 software, and a total of 5 pairs of primers were designed and optimized. , and carry out CY3 labeling on the 5' end tail sequence of the downstream primer. According to the designed primer interval range, select the highly variable interval of each representative strain to design the probe. In order to improve the influence of the steric hindrance of the probe, 15 T bases within 15 can be added at the 5' end, and the designed probe BLAST was performed to check its specificity. The primers and probes are shown in Table 1 and Table 2.
[0042] Table 1 Specific primer sequences
[0043]
[0044] Table 2 Specific probe sequences
[0045]
Embodiment 2
[0046] Embodiment 2 PCR amplification
[0047] (1) PCR amplification
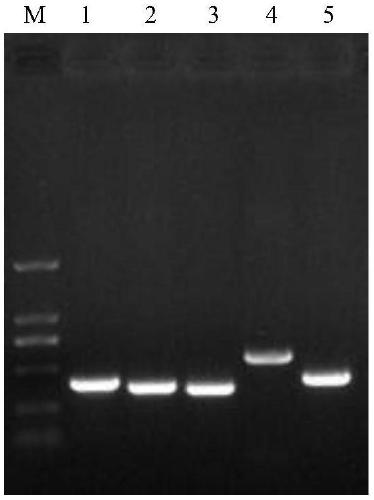
[0048] CAV1, CAV2, and CCV refer to the gene sequences of each virus registered on the NCBI website and synthesize plasmids by Shanghai Sangong Co., Ltd. CDV and CPV are extracted from clinically positive disease materials and passed plasmid mini-extraction reagents produced by Tiangen Biochemical Technology Co., Ltd. Cassette extraction to prepare plasmids. Using the extracted DNA and cDNA as templates, amplify by PCR. The PCR system is 25 μl: 2×ES TaqMaster Mix 12.5 μl, upstream primer (10 pmol / L) 1.0 μl, downstream primer (10 pmol / L) 1.5 μl, template 1.0 μl, make up the volume to 25 μl with RNase-Free Water. The PCR program was: pre-denaturation at 94°C for 3 min, denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min 15 s, a total of 35 cycles; extension at 72°C for 10 min.
[0049] The results of PCR amplification were as figure 1 shown.
[0050] (2) Construction of...
Embodiment 3
[0052] The establishment of the detection method of embodiment 3 gene chip
[0053] (1) Extract the nucleic acid of the sample to be tested and carry out reverse transcription;
[0054] (2) Amplify according to the method described in Example 2
[0055] (3) Chip preparation
[0056] Dilute the oligonucleotide probes synthesized by Shanghai Sangon Bioengineering Co., Ltd. to 40 μmol / L with distilled water, add 5 μL each to the A384 plate, and take 5 μL chip spotting solution for dilution, and use the Personal Arrayer 16 chip spot The sample instrument spots the probes on the aldehyde-based glass substrate (see Table 3), and repeats the spotting of each probe 3 times. The microarray layout includes the spotting quality control probe HEX, the positive quality control probe P , sample solution N for negative quality control probes, and 11 specific probes. After spotting the chip, place the chip at 37°C for more than 12h, then wash it with 0.2% SDS, wash it with clean water, sea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com