Disaccharide degrading enzyme gene and application thereof
A technology of gene and trehalase, which is applied in the field of biological enzymes, can solve the problems of no related reports on the application, and achieve the effects of increasing alcohol production, reducing residues, and reducing processing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
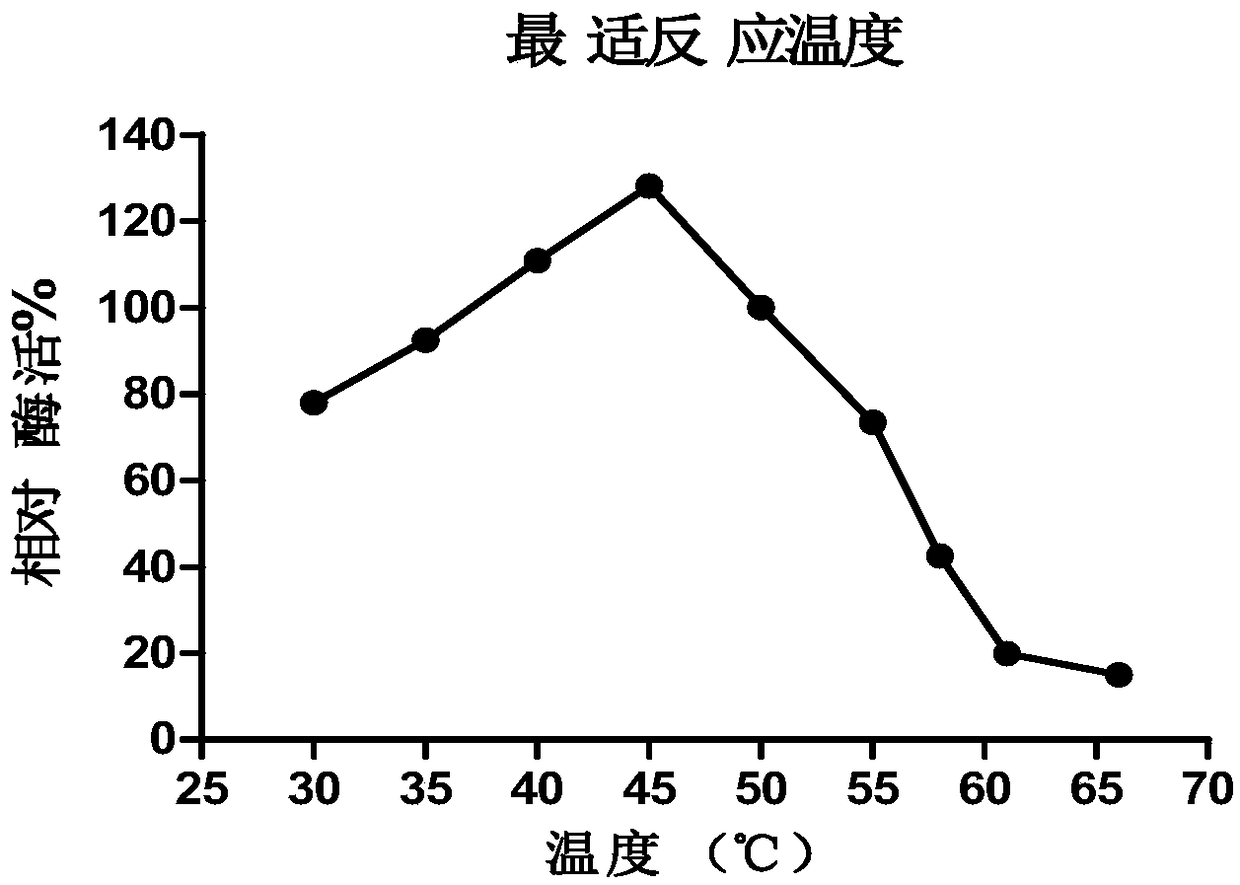
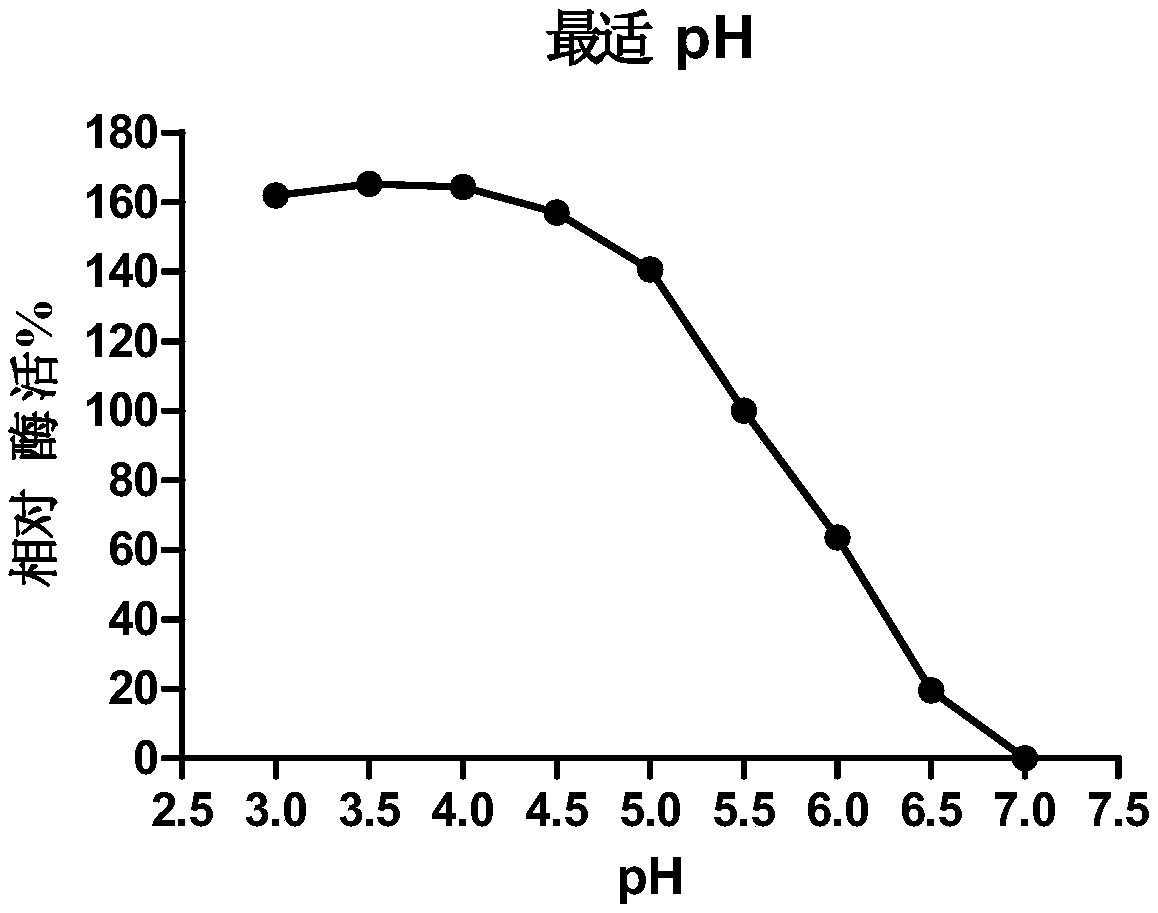
Image
Examples
Embodiment 1
[0038] Cloning of embodiment 1 trehalase gene TH3155
[0039] 1. Experimental materials and reagents:
[0040] 1. Strains and vectors
[0041] Escherichia coli Topl0, Pichia pastoris X33, Aspergillus niger 319 strain, plasmid pPICzαA, plasmid PBC-gla3, and Zeocin antibiotics were all purchased from Invitrogen.
[0042] 2. Gene
[0043] The strain Trichoderma reesei was purchased from the Guangdong Microorganism Culture Collection Center, and a trehalase gene TH3155 was amplified from the genome of the strain.
[0044] 3. Enzymes and reagents
[0045] Ultra-fidelity 2×Master Mix PCR polymerase was purchased from NEB Company; universal DNA Purification Kit, TIANprep Mini Plasmid Kit, and restriction endonuclease were purchased from Shanghai Sangon Company.
[0046] 4. Medium
[0047] The culture medium for Escherichia coli was LB medium (1% peptone, 0.5% yeast extract, 1% NaCl, pH7.0). LB+Amp medium is LB medium with ampicillin at a final concentration of 100ug / mL added....
Embodiment 2
[0062] Example 2 Pichia pastoris expression vector and host containing trehalase gene TH3155
[0063] Expression of trehalase gene TH3155 in Pichia pastoris: Digest the pPICzαA-TH3155 recombinant expression vector into linearized DNA fragments with Pme I restriction enzyme, purify the DNA fragments, and transform the linear pPICzαA-TH3155 DNA fragments by electroporation In the expression host Pichia pastoris X33 competent cells, spread on the YPD+Zeocin plate, culture upside down at 30°C for 3-4 days, pick a single colony into 2ml liquid YPD medium, culture at 30°C, 200rpm for 24 hours, 1% methanol was used to induce culture for 24 hours, the fermentation broth was collected, centrifuged, and the supernatant was taken as the crude enzyme solution.
Embodiment 3
[0064] Example 3 Aspergillus niger expression vector and host containing trehalase gene TH3155
[0065] Expression of trehalase gene TH3155 in Aspergillus niger: construct trehalase gene TH3155 in pPICzαA-TH3155 into Aspergillus niger expression vector PBC-gla3 with BglII restriction endonuclease and PmeI restriction endonuclease, transform Aspergillus niger 319 strain protoplasts. Coated on TZ + In the hygromycin plate, culture upside down at 32°C for 5-6 days, pick a single colony into 20mL liquid maltodextrin medium, culture at 32°C, 200rpm, and screen to obtain recombinant Aspergillus niger expressing trehalase.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com