Abelmoschus esculentus chloroplast microsatellite polymorphic marker primers and method for molecular identification of varieties
A technology of microsatellite polymorphism and molecular identification of varieties, applied in the field of molecular biology, can solve the problems of incomplete identification of okra resources, etc., and achieve the effects of multiple copies, small molecular weight, accurate identification and identification, and simple structure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1: okra chloroplast microsatellite molecular marker polymorphic primer (cpSSR) acquisition:
[0063] (1) Chloroplast microsatellite molecular marker sequence screening
[0064] The complete sequence of the chloroplast genome of okra (Abelmoschus esculentus) (NC_035234) was downloaded from the GenBank database (https: / / www.ncbi.nlm.nih.gov / ), the complete sequence of the chloroplast genome of okra was analyzed, and the molecular sequences of chloroplast microsatellites were screened. Use the MISA perl script software (http: / / pgrc.ipk-gatersleben.de / misa / ) to search for SSR sites (simple sequence repeats, SSR) distributed in the okra chloroplast genome, and the parameters are set as follows: single nucleotide repeat sequence (mononucleotide), repeating units greater than or equal to 10; dinucleotide repeating sequences (dinucleotides), repeating units greater than or equal to 5; trinucleotide repeating sequences (trinucleotides), repeating units greater than or...
Embodiment 2
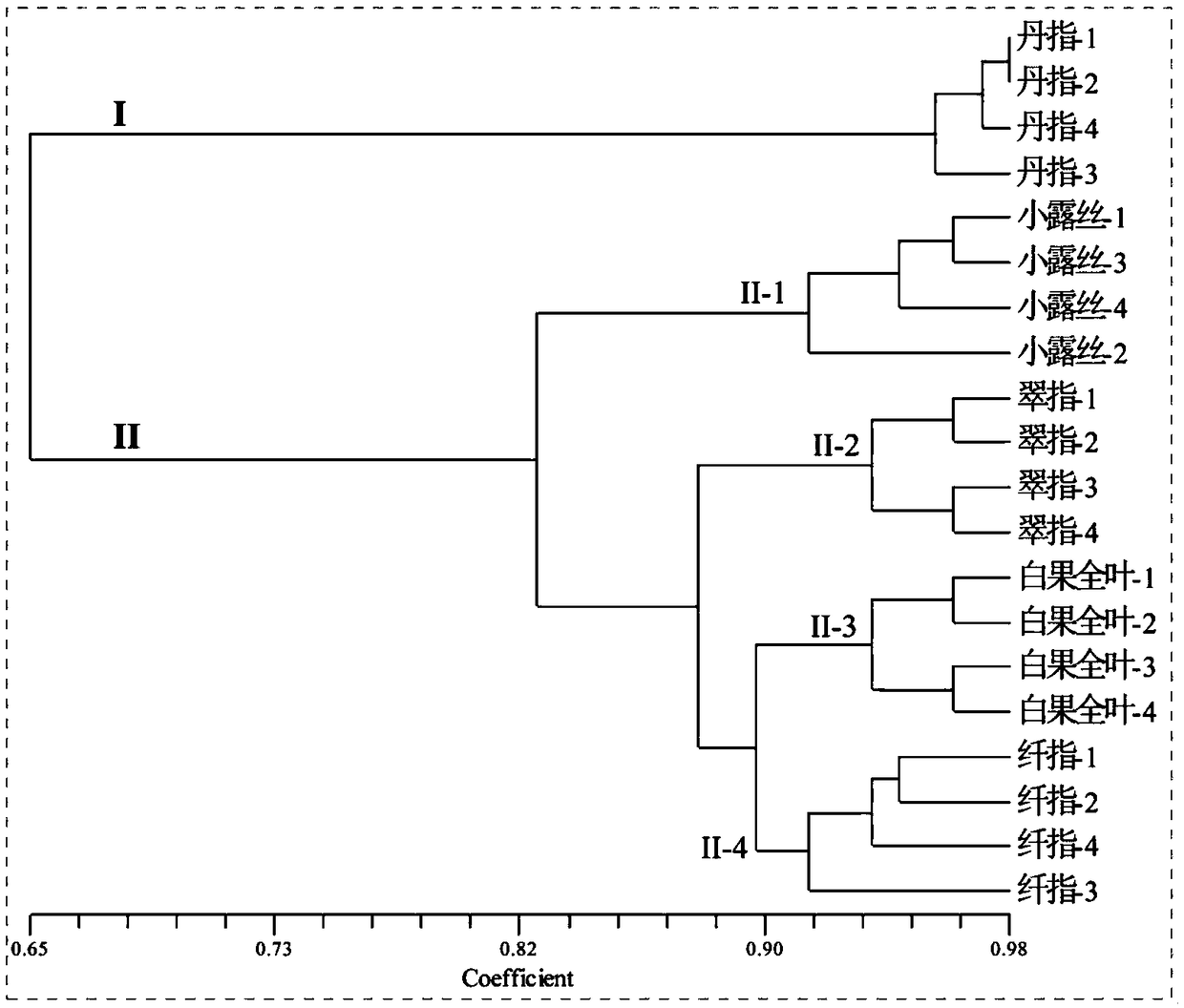
[0094] Embodiment 2: the present embodiment provides a kind of method that utilizes the polymorphic primer of the chloroplast microsatellite molecular marker that embodiment 1 provides to carry out the kinship analysis of okra variety, specifically as follows:
[0095] Sample: 20 parts of sample materials of 5 okra varieties provided by the embodiment of the present invention 1, and extract 20 parts of total genomic DNA of test samples, to detect the polymorphic primers of okra chloroplast microsatellite molecular markers in different varieties of okra versatility.
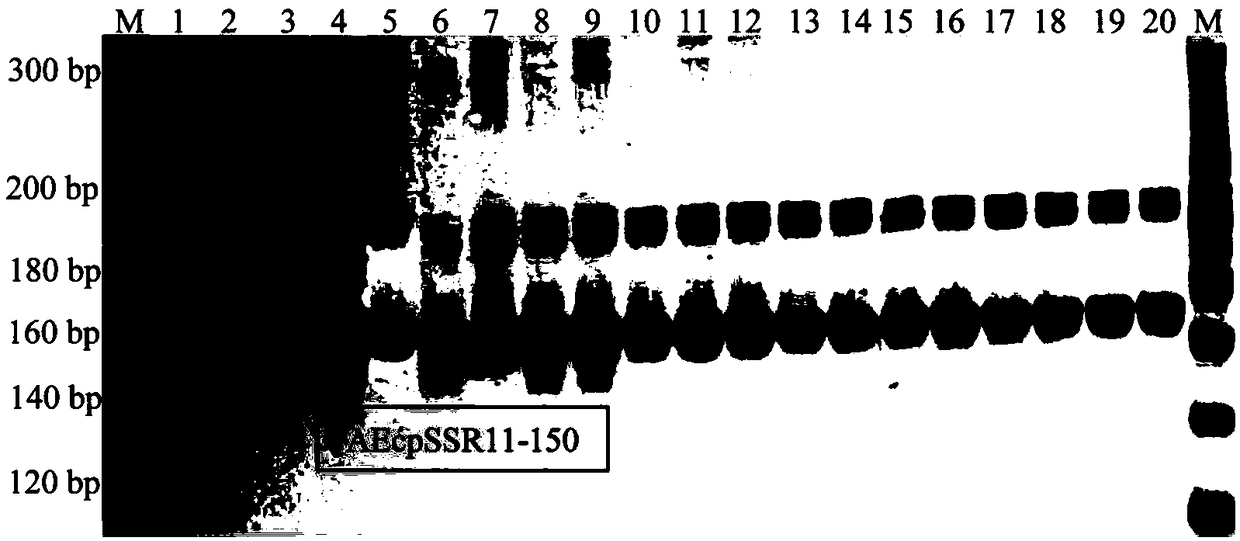
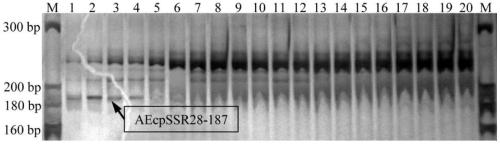
[0096] Using the polymorphic primers of chloroplast microsatellite molecular markers in okra provided in Example 1 as PCR extension primers, PCR amplification was performed on the total genomic DNA of 20 samples. Separation of the amplified product by non-denaturing polyacrylamide coagulation electrophoresis, if there is a DNA electrophoresis band at the same site, it is recorded as "1", if there is no DNA electro...
Embodiment 3
[0100] Embodiment 3: This embodiment provides a method for molecular identification of okra varieties using the polymorphic primers of chloroplast microsatellite molecular markers provided in embodiment 1. details as follows:
[0101] (1) extract the total genomic DNA of the sample of the tested okra variety;
[0102] (2) With the DNA extracted in step (1) as a template, the polymorphic primers of the okra chloroplast microsatellite molecular markers provided by Example 1 are used as PCR amplification primers to carry out PCR amplification respectively;
[0103] (3) Carrying out 8% non-denaturing polypropylene gel electrophoresis detection to the PCR product of step (2), obtaining the DNA fingerprint of the sample to be tested;
[0104] (4) Carry out band type statistics on the DNA fingerprint of the sample in step (3), and determine the okra species to be tested by comparing and judging specific electrophoretic bands.
[0105] It was found that 4 pairs of cpSSR primers ampl...
PUM
| Property | Measurement | Unit |
|---|---|---|
| quality score | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com