A combination of primers and probes for the detection of Fusarium solani rot based on rpa-lateral flow chromatography and its application
A technology for Fusarium solani and lateral flow chromatography, which is applied in the biological field, can solve problems such as no relevant application reports, and achieves good amplification effect, increased sensitivity and good specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] The genomic DNA of the tested pathogenic bacteria strain was extracted, and the specific extraction process was as follows:
[0033] Take a small amount of mycelium powder, add 900 μL 2% CTAB extract and 90 μL 10% SDS, vortex and mix well, put in a water bath at 60°C for 1 hour, and invert several times every 10 minutes. Centrifuge at 12000rpm for 10min, take the supernatant and add an equal volume of phenol / chloroform / isoamyl alcohol (25:24:1), mix by inversion, and centrifuge at 12000rpm for 10min; transfer the supernatant to a new tube, add an equal volume of chloroform, Gently invert to mix and centrifuge at 12000rpm for 5min. Take the supernatant and transfer it to a new tube, add 2 times the volume of absolute ethanol and 1 / 10 volume of 3M NaAc (pH5.2), and precipitate at -20°C (>1h). Centrifuge at 12000rpm for 10min, pour off the supernatant, wash the precipitate twice with 70% ethanol, and dry at room temperature. Add an appropriate amount of sterilized ultrap...
Embodiment 2
[0035] Using the extracted DNA as a template and using the designed RPA primers, perform the RPA reaction in the following reaction system:
[0036] Sample detection: Add 29.5 μL of buffer buffer, 2.1 μL of 10 μM upstream primer, 2.1 μL of 10 μM downstream primer, and 0.6 μL of probe to a 0.2 mL TwistAmp reaction unit tube (TwistAmp Basickits, Twist) containing lyophilized enzyme powder , DNA 2.0μL, MgAc 2.5μL were added to the inside of the PCR tube cap, and deionized water was added to 50μL; the RPA amplification system was thoroughly mixed, centrifuged at 5,000×g for 10s, placed on a metal bath at 39°C for 30min, and incubated for 4 Minutes, the reaction tube was mixed again, centrifuged for 3-5 s, and placed in a water bath at 39°C to continue the reaction for 30 min.
[0037] Wherein, the length of the RPA primer is generally 30 to 35 nucleotides. Too short primers will seriously affect the activity of the recombinase. Long primers do not necessarily improve the amplific...
Embodiment 3
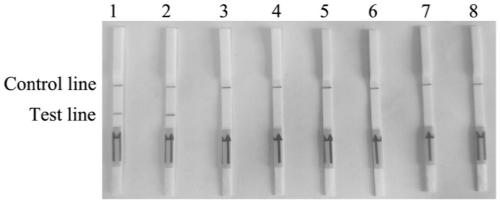
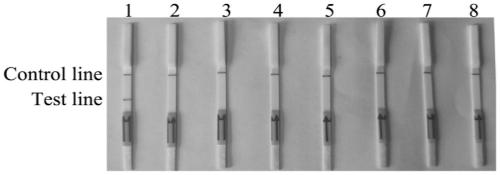
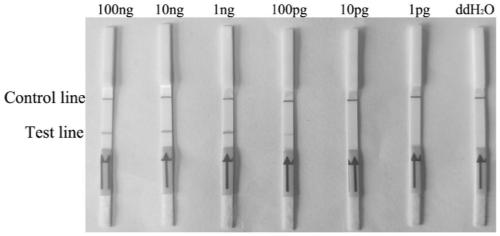
[0042] In order to verify the specificity of the RPA lateral flow chromatography test strip detection method, Fusarium solani solani and other Fusarium and pathogenic bacteria were used as test materials (Table 1), and the RPA lateral flow chromatography test strip detection method The results show that there are two brown bands on the test strip of Fusarium solani rot, one is in the quality control area and the other is in the detection area, the result is positive. The test strips of other Fusarium solani and pathogenic bacteria only appear in the quality control area A brown band with no band in the test area is negative, indicating that Fusarium solani solani is not present in the sample.
[0043] Select different species of Fusarium solani rot (Fusarium solani rot; Fusarium oxysporum; Fusarium graminearum; Fusarium moniliforme; Fusarium equiseti; Pythium ultima; Anthracnose flathead; Phytophthora sojae; Verticillium dahliae; Rhizoctonia solani; Magnaporthe oryzae, etc.) w...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com