Corynespora cassiicola succinodehydrogenase subunit c N75S resistance mutation detection primer and detection method
A technology of succinate dehydrogenase and multi-main Corynebacterium, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve the problems of undetected resistance mutations, etc., and achieve easy promotion, use, and detection Strong specificity and simple instrument requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
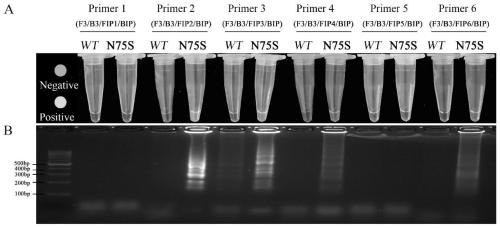
[0051] Example 1: Establishment of a method for isothermal detection of the N75S resistance mutation loop mediated by the c subunit of the succinate dehydrogenase of Corynespora polymorpha
[0052] The Corynespora leaf spot strain containing the N75S resistance mutation was used as the test object, and the wild-type sensitive strain was used as the negative control.
[0053] 1. Primer design:
[0054] Based on the nucleic acid sequence of the c subunit of the succinate dehydrogenase of Corynespora polyprimus with N75S resistance, four sets of specific primers were designed using the LAMP primer design software PrimerExplorer V4, and one set of primers was screened out according to the principle of LAMP primer design and artificial alkali Base pair mismatch (shown underlined), the primer sequence is as follows (5'-3'):
[0055] The first set of primers:
[0056] F3: CGCCCGCAGATCACCT; as shown in SEQ ID No. 1;
[0057] B3: TGGCCGCAACCTTCGC; as shown in SEQ ID No. 2;
[0058] FIP1: CGAAGAGG...
Embodiment 2
[0097] Example 2: Optimization of LAMP detection system
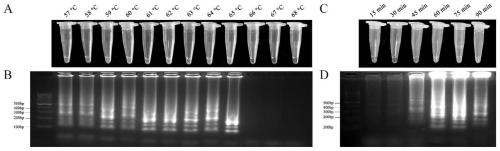
[0098] According to Example 1, the best primer can be selected as the second set of primers. Therefore, through various factors affecting the construction system, including inner and outer primer concentrations (inner primer 0.8-2.0 μM, outer primer 0.1-0.6 μM), betaine Concentration (0.8-1.6M), Mg 2+ Concentration (1.0-7.0mM), Bst DNA polymerase content (1-6U), dNTPs concentration (0.7-1.3mM), temperature (57-68℃) and time (15-90min) to screen and determine the optimal detection system . Reaction system (10μL).
[0099] The LAMP results were analyzed by two methods: direct visual observation after adding 10000×Sybr Green I and 3% agarose gel electrophoresis.
[0100] The best ratio is 1×ThermoPol buffer (20mM Tris-HCl, 10mM KCl, 2mM MgSO 4 , 10mM(NH 4 ) 2 SO 4 , 0.1% TritonX-100), F3 0.2μM, B3 0.2μM, FIP2 1.6μM, BIP 1.6μM, dNTPs 1.0mM, MgSO 4 6mM, Bst DNA polymerase 2U, template DNA 0.5μL, betaine 0.8M. by figure 2 It ca...
Embodiment 3
[0101] Example 3: Specificity of LAMP detection method in the present invention
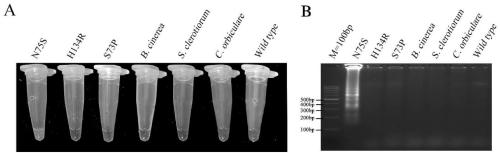
[0102] According to the optimal reaction system and reaction conditions of the above-mentioned Example 2, at the same time, it can be used for the N75S, H134R, S73P mutant Corynespora polysporum, B. cinerea, S. clerotiorum, and Cucumber anthracnose. (C.orbiculare) and wild type strains (wild type) genomic DNA as templates for detection, to test the specificity of the LAMP method.
[0103] The LAMP results were analyzed by two methods: direct visual observation after adding 10000×Sybr Green I and 3% agarose gel electrophoresis. by image 3 It can be seen that only specific yellow and diffuse electrophoretic bands appeared in the N75S reaction tube, while the other reaction tubes kept the dye orange and no diffuse electrophoretic bands. The results show that the LAMP method of the present invention has strong specificity, can specifically detect N75S, and has no cross-reactivity with other mutations on...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com