Zymomonas mobilis genome editing method based on CRISPR-Cas12a system and application of method
A Zymomonas, genome editing technology, applied in the field of gene editing, can solve the problems of restricting research and application, large workload, long time consumption, etc., and achieves the effect of wide application range, simple operation and simple process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Construction of an inducible gene editing system suitable for Zymomonas mobilis
[0040] The present invention uses Zymomonas mobilis ZM4 as a model strain, and realizes directional editing of the genome by constructing an inducible gene editing system suitable for Zymomonas mobilis. The specific experimental process is as follows:
[0041] 1. Construction of an inducible Cas12a recombinant strain
[0042] In the present invention, firstly, the nuclease Cas12a derived from Francisella novicida was integrated into the ZMO0038 site in the Z.mobilis ZM4 genome by the method of homologous recombination, and an inducible promoter was used to control the expression of the nuclease. Recombinant strain ZM-Cas12a.
[0043] The specific construction process is as follows:
[0044] (1) Construction of recombinant plasmid
[0045]The Cas12a gene sequence, the resistance selection marker (spectinomycin), the inducible promoter gene sequence (tetracycline-inducible promo...
Embodiment 2
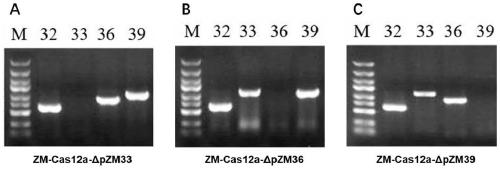
[0073] Example 2 Application of Zymomonas mobilis genome editing method based on CRISPR-Cas12a system in elimination of endogenous plasmids
[0074] 1. Selection of target sites
[0075] The genomic data of Z. mobilis ZM4 has been published, which contains 4 endogenous plasmids and named according to the size of the sequences as pZM32 (32,791bp), pZM33 (33,006bp), pZM36 (36,494bp) and pZM39 (39,266bp) . Sequence analysis showed that the four endogenous plasmid strains edit the replicase. If the replicase is inactivated, the endogenous plasmid will lose the ability to replicate and the endogenous plasmid will be eliminated from the strain.
[0076] The present invention selects a sequence of 23 bp downstream of the TTTN site of the PAM site from the replicase gene of the endogenous plasmid as the targeting guide sequence of the guide RNA in the construction of the target plasmid to guide the cleavage of the target site by the nuclease. The forward primer is 5'-AGAT+(target se...
Embodiment 3
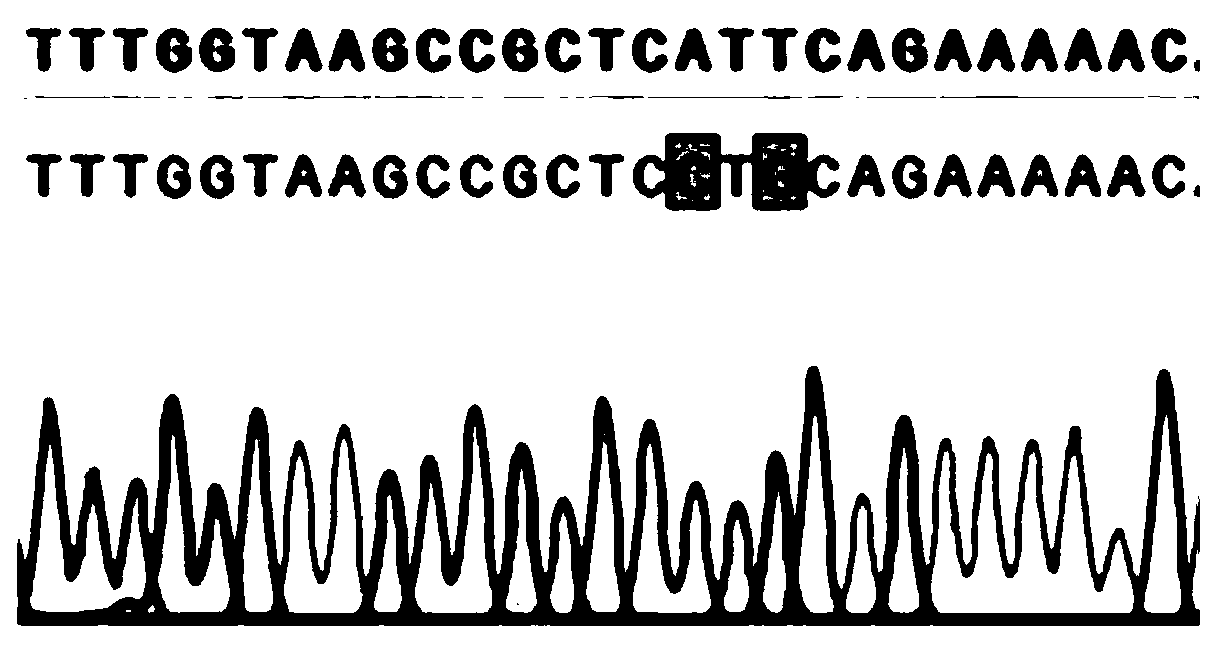
[0105] Example 3 Application of Zymomonas mobilis genome editing method based on CRISPR-Cas12a system in point mutation
[0106] 1. Selection of target sites
[0107] Select ZMO1237 in the Z. mobilis ZM4 genome as the target site, perform point mutation on it, and select the 23bp sequence downstream of the PAM site TTTN site from the target gene as the targeting guide sequence for constructing the guide RNA in the target plasmid, and guide the nuclease Cleavage of target sites. The forward primer is 5'-AGAT+(target sequence)-3', and the reverse primer is 5'-TGAC+(target sequence complementary sequence)-3'.
[0108] The sequence of the guide RNA primer is as follows, where the underlined part is complementary to the enzyme cutting site:
[0109] Ldh-F: AGAT GTAAGCCGCTCATTCAGAAAAAC, see SEQ ID NO: 15;
[0110] Ldh-R: TGAC GTTTTTCTGAATGAGCGGCTTAC, see SEQ ID NO:16.
[0111] 2. Construction of target plasmids
[0112] Ligate the guide RNA primer sequence to the editing pl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com