Method for rapidly and precisely detecting African swine fever virus on basis of CRISPR/Cas12a and application
A kind of African swine fever virus and precise technology, applied in the field of pathogen detection, can solve the problems of difficult and indeterminate negative and positive sample detection and accurate judgment, prevention and control, etc., achieve short detection time, facilitate early and rapid detection, and low cost Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Design of crRNA
[0044] The core of the CRISPR / Cas12a detection method lies in crRNA, so the quality of crRNA is directly related to the sensitivity and accuracy of the detection method. In order to screen the crRNA with the highest efficiency, we designed 19 ssDNAs against ASFV VP72 gene (GenBank accessionno.MH766894.1), and prepared 19 crRNAs (the sequences are shown in SEQ ID NO.1-19).
[0045] (1) Annealing system as shown in Table 1
[0046] Table 1
[0047]
[0048]
[0049] Add the above components to a 200 μl centrifuge tube, mix well and centrifuge. Put into a PCR machine for annealing reaction: 37°C, 30min; 95°C, 5min; drop 5°C every minute to 25°C to obtain Template DNA. ssDNA sense and ssDNA antisense were synthesized by Shanghai Shenggong.
[0050] (2) Transcription
[0051] with MEGAshortscript TM T7 Transcription Kit (Invitrogen, USA) was used for transcription, and the reaction system was shown in Table 2:
[0052] Table 2
[00...
Embodiment 2
[0057] Example 2 Establishment of CRISPR / Cas12a detection method for ASFV
[0058] 1. Amplification of the target fragment
[0059] (1) Extract viral DNA
[0060] Take 200 μL of serum samples and use RaPure Viral RNA / DNA Kit (Magen, China) to process according to the operation steps to extract viral nucleic acid.
[0061] (2) RPA amplification
[0062] The RPA-F / RPA-R designed for RPA amplification is shown in SEQ ID NO. 20-21.
[0063] The RPA amplification system is shown in Table 3
[0064] table 3
[0065]
[0066] Add 46 μL of the above mixture to a Twist Amp NFO kit (TwistDxTM, Cambridge, United Kingdom) reaction tube containing lyophilized enzyme powder, pipette the lyophilized enzyme powder until it is completely dissolved, and then add 4 μL of magnesium acetate solution to the reaction tube. , Mix well, and put the reaction tube into a 37°C constant temperature incubator for 10 min to complete the RPA amplification. The RPA amplification products were subject...
Embodiment 3
[0072] Example 3 Screening crRNA with the highest efficiency
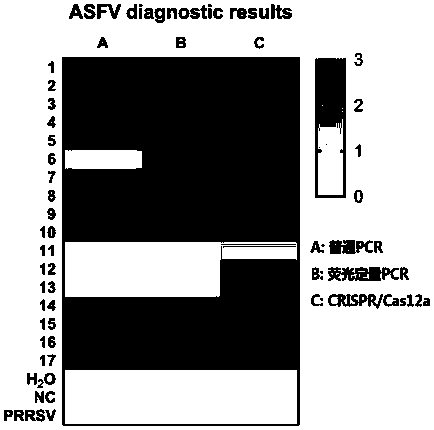
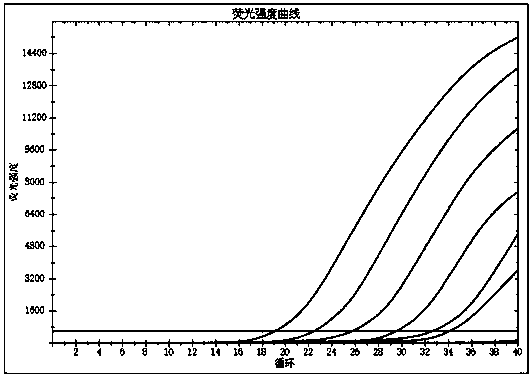
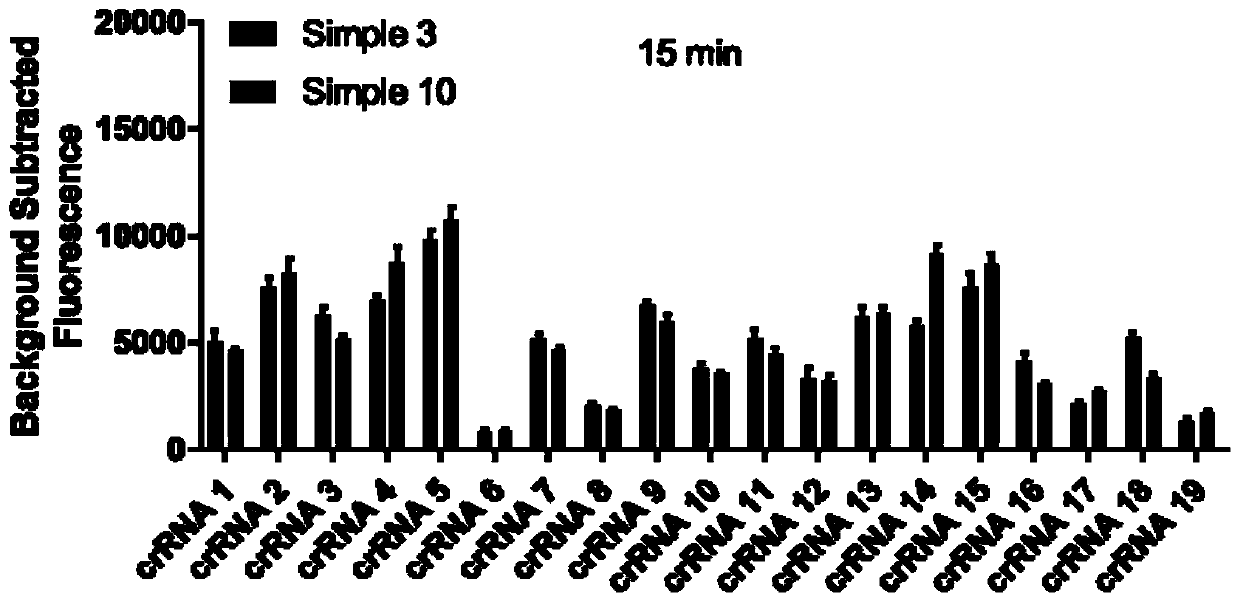
[0073] RaPure Viral RNA / DNA Kit (Magen, China) was used for 2 clinical samples of ASF virus nucleic acid positive samples, and the virus nucleic acid was extracted according to the operation steps. Then, according to the RPA amplification method and conditions in Example 1, RPA amplification was carried out. Finally, the reaction was carried out according to the CRISPR / Cas12a detection method and conditions of ASFV in Example 1. The 19 crRNAs prepared above were used as the crRNAs in the reaction system, and the reaction time was 15 or 30 min. After the reaction, the fluorescence value was read with a BioTek microplate reader.
[0074] The result is as figure 1 , figure 2 Shown: after the reaction time of 15 and 30 min, the fluorescence values of crRNA5 and crRNA15 are the highest, indicating that among the 19 crRNAs, these two have the best effect and the highest sensitivity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com