Method for identifying candida glabrata by high-resolution-ratio melting curve
A high-resolution melting, Candida glabrata technology, applied in microorganism-based methods, biochemical equipment and methods, and microbial determination/inspection, etc., can solve the problems of complex operation, misidentification, long time consumption, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] In the following examples, unless otherwise specified, all are conventional methods.
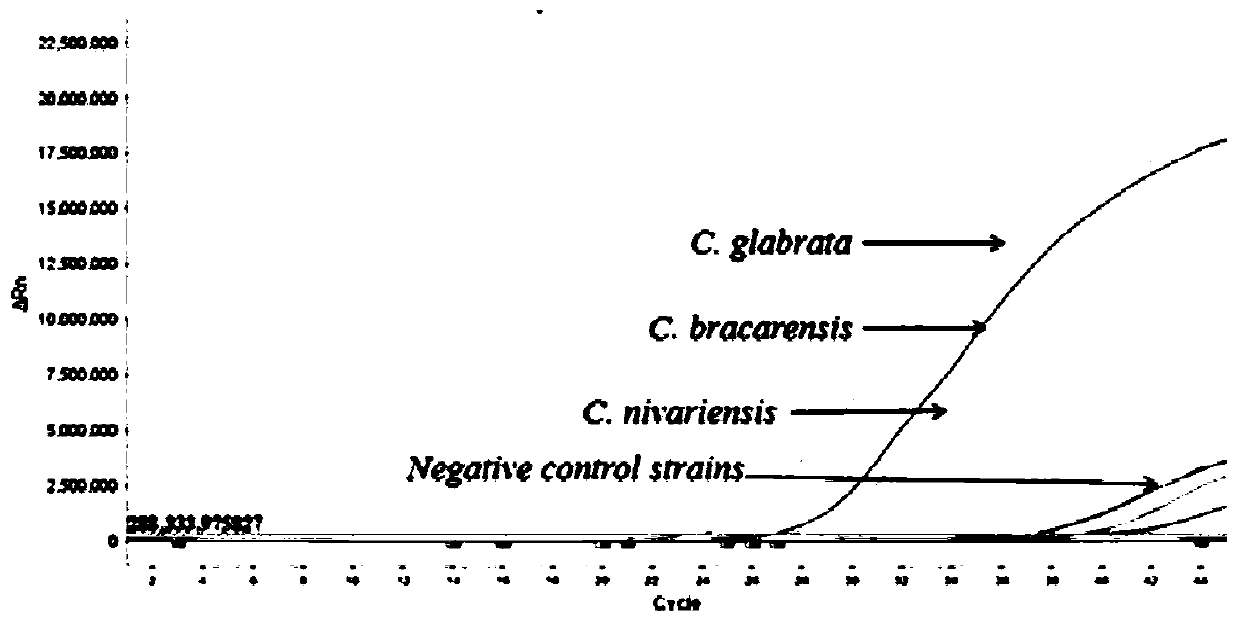
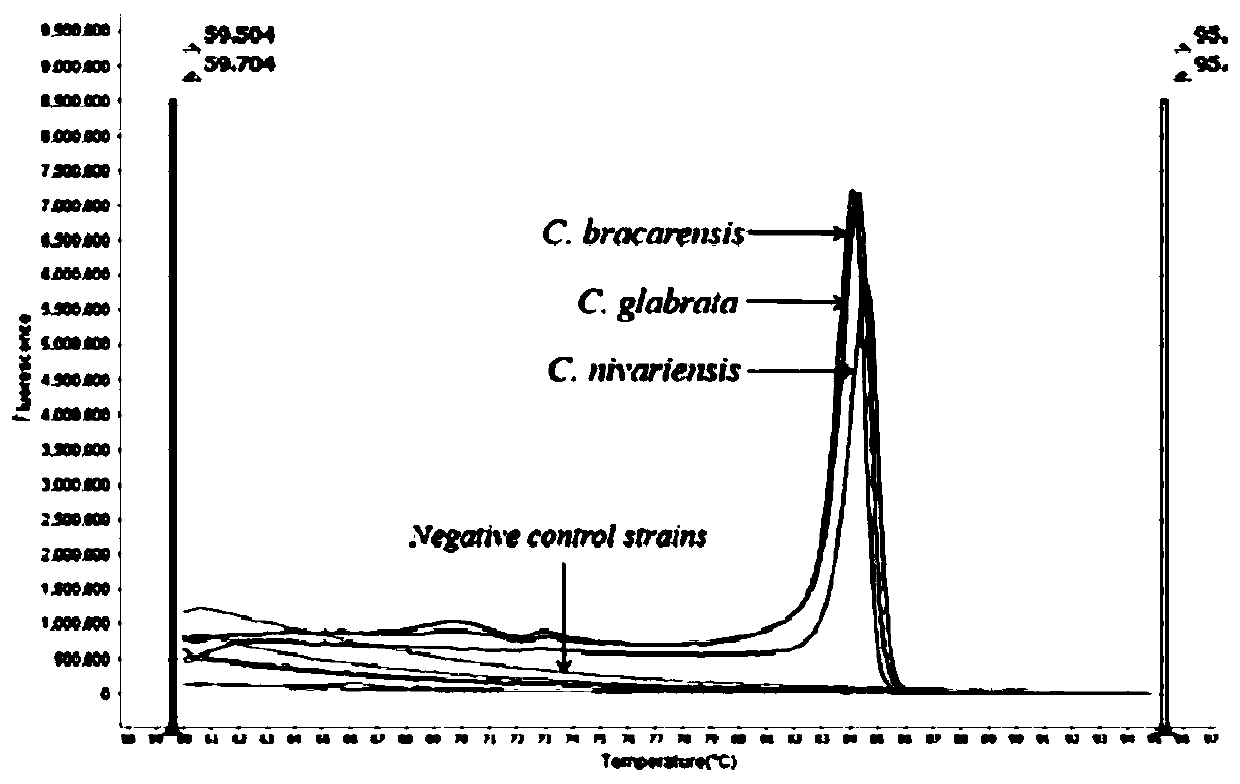
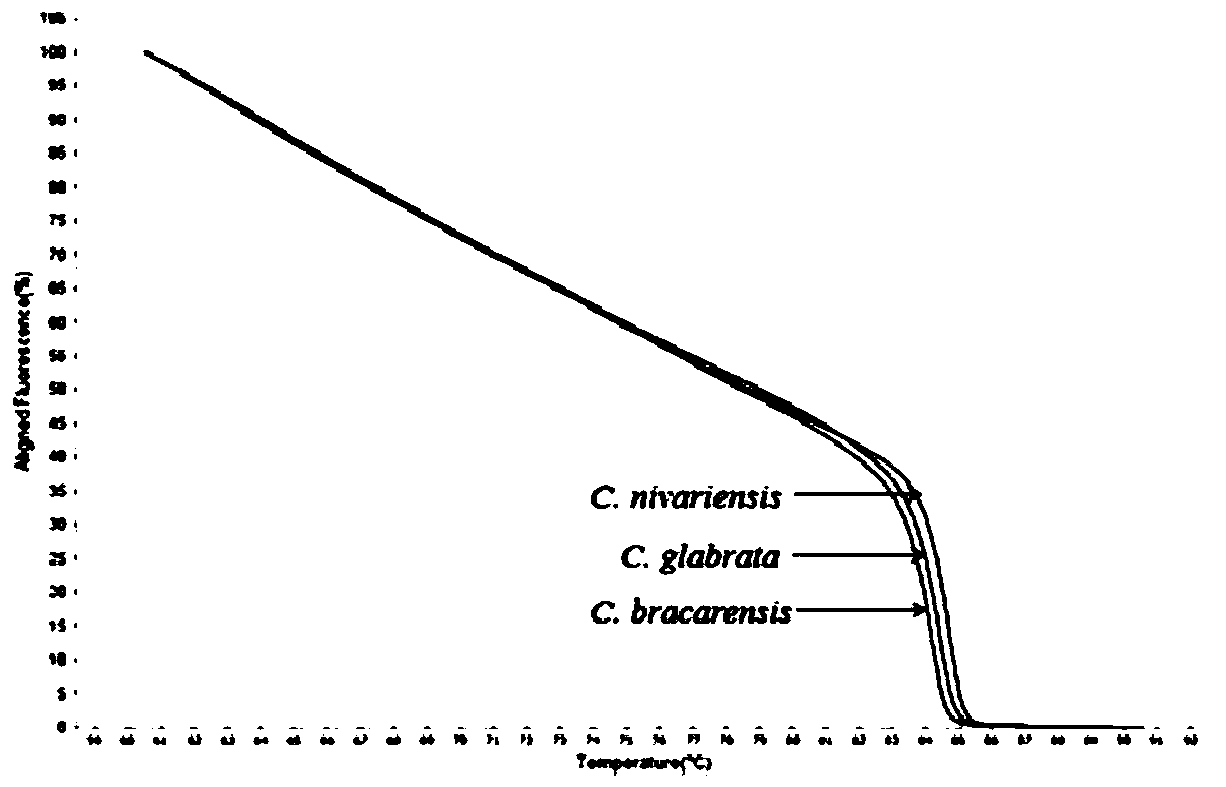
[0034] Experimental strains: Candida glabrata (C. glabrata) - Candida glabrata, Candida nivariensis (C. nivariensis) - Candida Nivaria, Candida bracarensis (C. bracarensis) - Candida bugara;
[0035]Control strains: Candida albicans-Candida albicans, Candida tropicalis-Candida tropicalis, Candida parapsilosis-Candida parapsilosis, Candida metapsilosis-Candida pseudopsilosis, Candidaorthopsilosis-Candida pseudopsilosis, Candida krusei-Candida krusei, Candida haemulonii - Candida haemulonii - Candida pseudohaemulonii - Candida pseudohaemulonii - Candida pseudohaemulonii - Candida haemuloniivar.vulnera - Candida haemuloniivar. - Aspergillus sydowii, Aspergillus sydowii, Aspergillus flavus, Aspergillus flavus, Alternaria alternate, Cladosporium sphaerospermum, Curvularia lunata, Fusarium solani fungus, Hortaea werneckii-Werneck phyllomus, Kodamaea ohmeri-Omer Kodak bacteria, Talaromycesf...
Embodiment 2
[0056] In the following examples, unless otherwise specified, all are conventional methods.
[0057] Experimental strains: Candida glabrata (C. glabrata) - Candida glabrata, Candida nivariensis (C. nivariensis) - Candida Nivaria, Candida bracarensis (C. bracarensis) - Candida bugara;
[0058] Control bacterial species: Select some bacterial species in Example 1 as the control bacterial strain: Candida albicans-Candida albicans, Candida tropicalis-Candida tropicalis, Candida parapsilosis-Candida parapsilosis, Candidametapsilosis-like Candida smoothis, Candida orthopsilosis-Pseudomonas Candida smooth, Candida krusei - Candida krusei, Candida haemulonii - Candida hemolongii, Candida duobushaemulonii - Candida pseudohemulonii, Candida haemulonii var. vulnera - Candida haemulonii var. Candida auris, Candida auris - Candida auris.
[0059] Both the experimental and control strains were obtained from the China Hospital Invasive Mycosis Surveillance Network (CHIF-NET). The total DNA...
Embodiment 3
[0070] In the following examples, unless otherwise specified, all are conventional methods.
[0071]Experimental strains: Candida glabrata (C. glabrata) - Candida glabrata, Candida nivariensis (C. nivariensis) - Candida Nivaria, Candida bracarensis (C. bracarensis) - Candida bugara;
[0072] Control bacterial species: select some bacterial species in Example 1 as the control bacterial strain: Trichosporon asahii-Asahi Trichosporon, Aspergillus sydowii-poly Aspergillus, Aspergillus flavus-Aspergillus flavus, Alternariaalternate-Alternaria, Cladosporium sphaerospermum- Cladosporium cocciformis, Curvularia lunata- Curvularia lunata, Fusarium solani- Fusarium solani, Hortaea werneckii- Wernecke exophyllum, Kodamaeaohmeri- Omer Kodak, Talaromyces funiculosus- rope-shaped basket fungus, Trichophyton rubrum- Trichophyton rubrum.
[0073] All the other steps are the same as in Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com