A gene for regulating the formation of tea leaves and its application
A technology of tea hair and gene, which is applied in the field of tea tree MYB transcription factor gene and its encoded protein, can solve the problems of unclear molecular mechanism, hinder the in-depth understanding of tea tree development biology and tea quality formation mechanism, and achieve rich understanding know the effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1. Cloning and sequence structure analysis of CsMYB184 gene
[0018] Shucha, a national-level fine variety of tea tree, was planted in the Agricultural Industrial Park of Anhui Agricultural University, Hefei, Luyang District, Anhui Province, and the young leaves were used for RNA extraction. The total RNA was extracted with Trizol reagent (Invitrogen, USA) according to the instructions, and the RNA content and quality were detected with a spectrophotometer. Reverse transcription to generate the first strand: take 1 μg of RNA as a template, and synthesize cDNA according to the instructions of Promega M-MLV reverse transcriptase supporting reagent. After optimization, take an appropriate amount of reverse transcription product for subsequent PCR. The CsMYB184 gene was amplified by conventional PCR using cDNA as RT-PCR template. Upstream primer: (5'-ATGGCTCCGAAGAGCAGTGA-3'), downstream primer: (5'-TTACCATTTATCGGTAAGTGCC-3'). The 25 μL PCR reaction system was: 10...
Embodiment 2
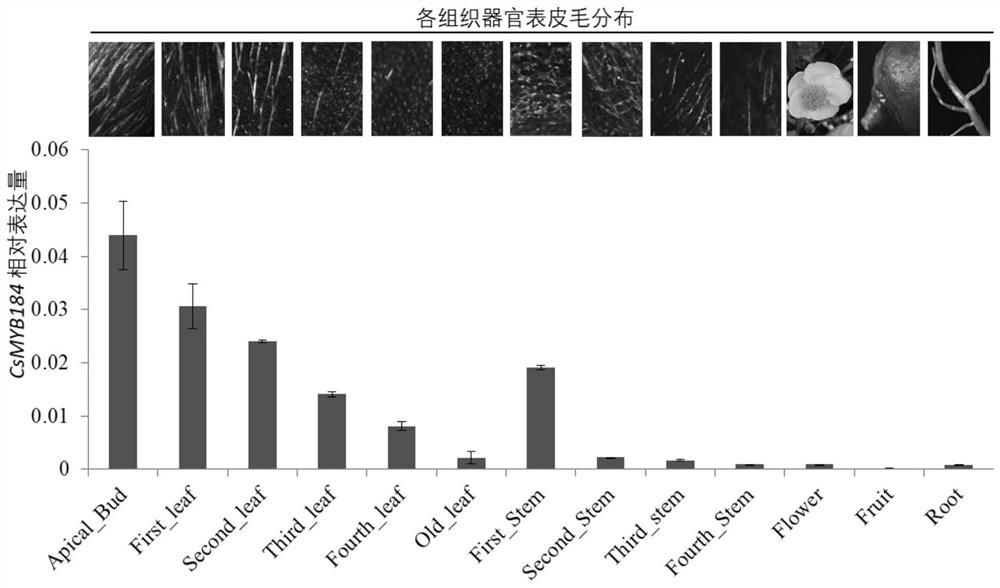
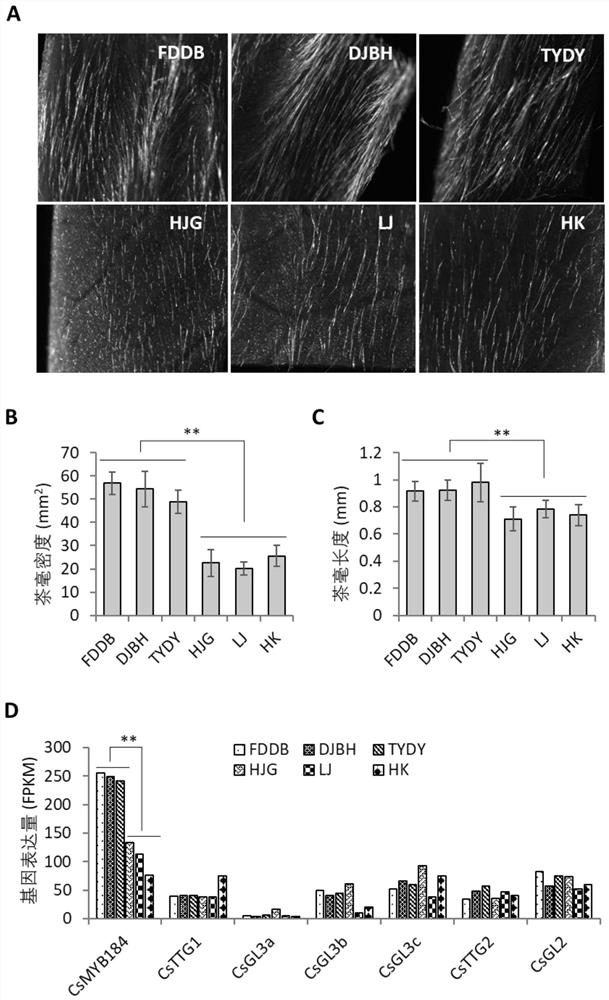
[0023] Example 2. The correlation between the expression difference of CsMYB184 gene and the difference of tea millet distribution:
[0024] 1. The distribution and CsMYB184 gene expression in different tissues of tea plant
[0025] The national-level tea tree variety, Fuding Dabai, was planted in the Agricultural Industrial Park of Anhui Agricultural University, Hefei, Luyang District, Anhui Province. 13 tissues and organs were used to analyze the distribution and gene expression of tea leaves. The 13 tissues and organs include young shoots, one leaf, two leaves, three leaves, four leaves, old leaves, the first stem segment, the second stem segment, the third stem segment, the fourth stem segment, flowers, fruits, and roots. The distribution of tea hair of 13 varieties was photographed and recorded by stereo microscope or digital camera. These samples were also used for total RNA extraction and cDNA first-strand synthesis. The reverse transcription product (the first strand...
Embodiment 3
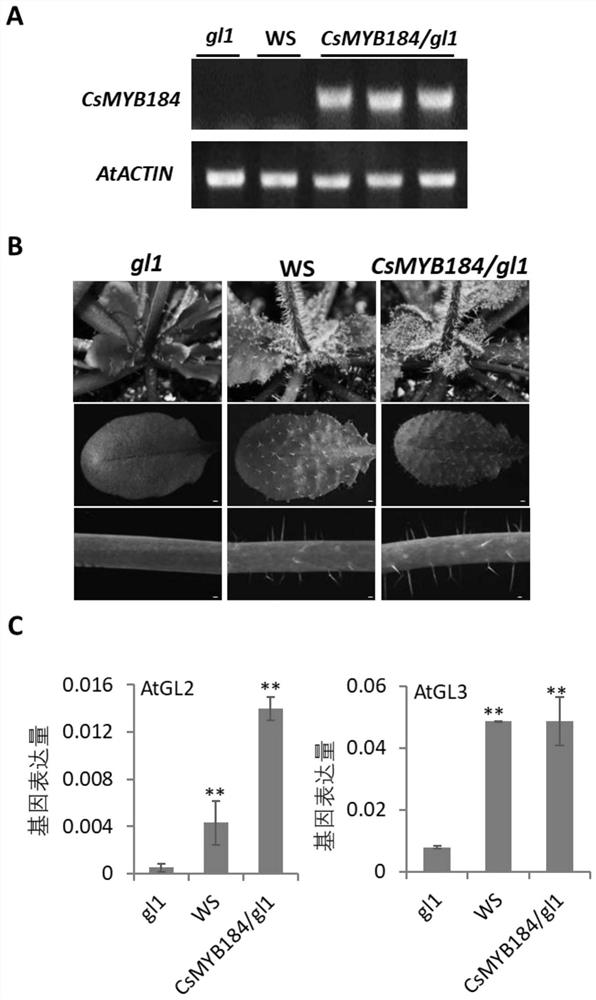
[0030] Example 3. Functional verification of CsMYB184 gene in Arabidopsis thaliana
[0031] 1. Construction of CsMYB184-pB2GW7 vector
[0032] Using the pMDTM 19-T Simple::CsMYB184 plasmid as a template, PCR amplification was performed with the following primers:
[0033] Upstream primer: (5'-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCTCCGAAGAGCAGTGA-3'),
[0034] Downstream primer: (5'-GGGGACCACTTTGTACAAGAAAGCTGGGTTTACCATTTATCGGTAAGTGCC-3'),
[0035] PCR products were recovered with 1% agarose gel electrophoresis bands. Using Gateway cloning technology, 1 μl of PCR recovery product was added, pDONR221 intermediate vector of equivalent quality was added, and finally 1 μL of BP Clonase Mix was added, and after overnight at room temperature, DH5α was transformed and sent for sequencing. The plasmids of the correct positive clones were sequenced, 1 μL of plasmid was taken and an equal amount of pB2GW7 overexpression vector was added. Finally, 1 μL of LR Clonase Mix was added. After o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com