Plant genome directional base editing skeleton vector and application thereof
A plant genome and base editing technology, which is applied in the field of plant genome directed base editing backbone vectors, can solve the problems of low editing efficiency and limited editing window, and achieve the goal of expanding the base editing window, good application prospects, and efficient directed editing Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
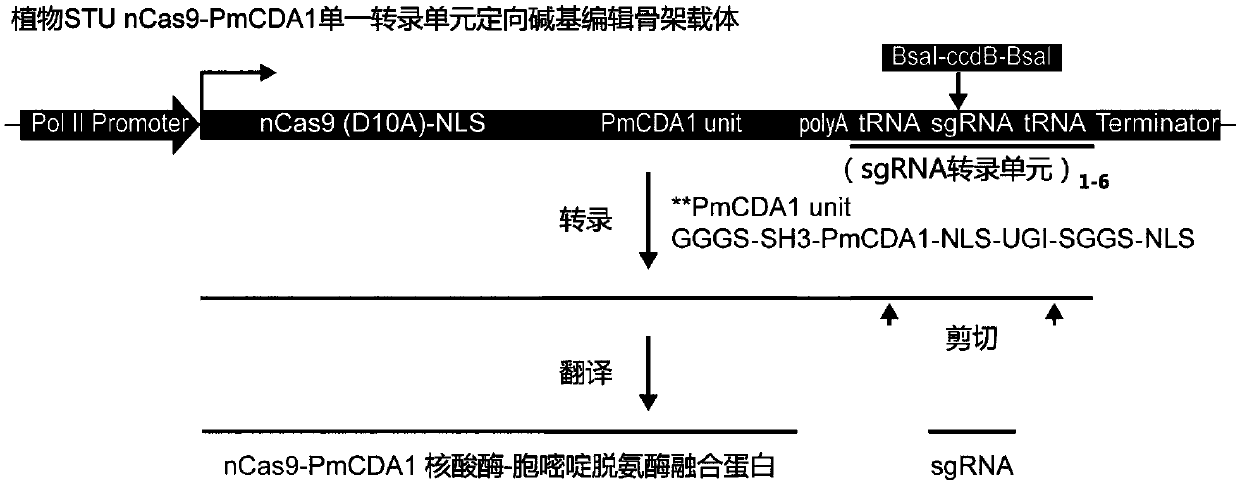
[0064] Example 1 Construction of Plant STU nCas9-PmCDA1 Single Transcription Unit Directed Base Editing Backbone Vector
[0065] The present invention designs a STU nCas9-PmCDA1 single transcription unit directional base editing backbone carrier for plant genome engineering, and its core unit is pZmUbi1-nCas9 ORF-PmCDA1-Poly A-sgRNA cloning scaffold from 5' to 3' direction -HSP T. Among them: pZmUbi1 is the maize pZmUbi1 promoter (the basic STU nCas9-PmCDA1 single transcription unit-directed base-editing backbone carrier scheme can be used to realize the replacement of different PolII-type promoters through AscI and SbfI double enzyme digestion); nCas9 ORF is the suppurative chain Cocci (Streptococcus pyogenes) nuclease protein D10A mutant coding frame (including C-terminal NLS signal peptide); region, NLS signal peptide, UGI coding region, SGGS linker, NLS signal peptide); Poly A is the poly A region; sgRNA cloning scaffold (abbreviated as sgRNA CS) is the sgRNA clone and tr...
Embodiment 2
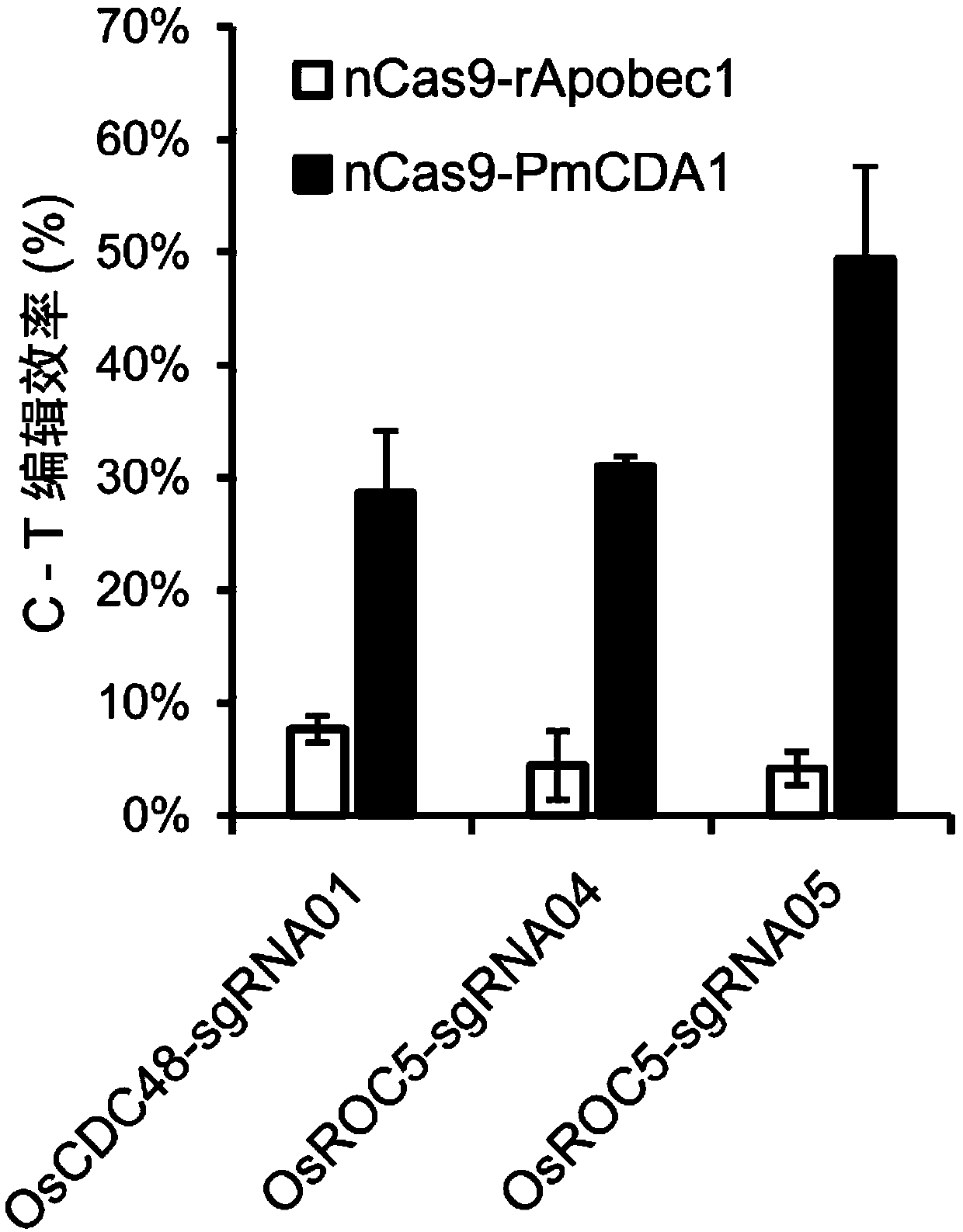
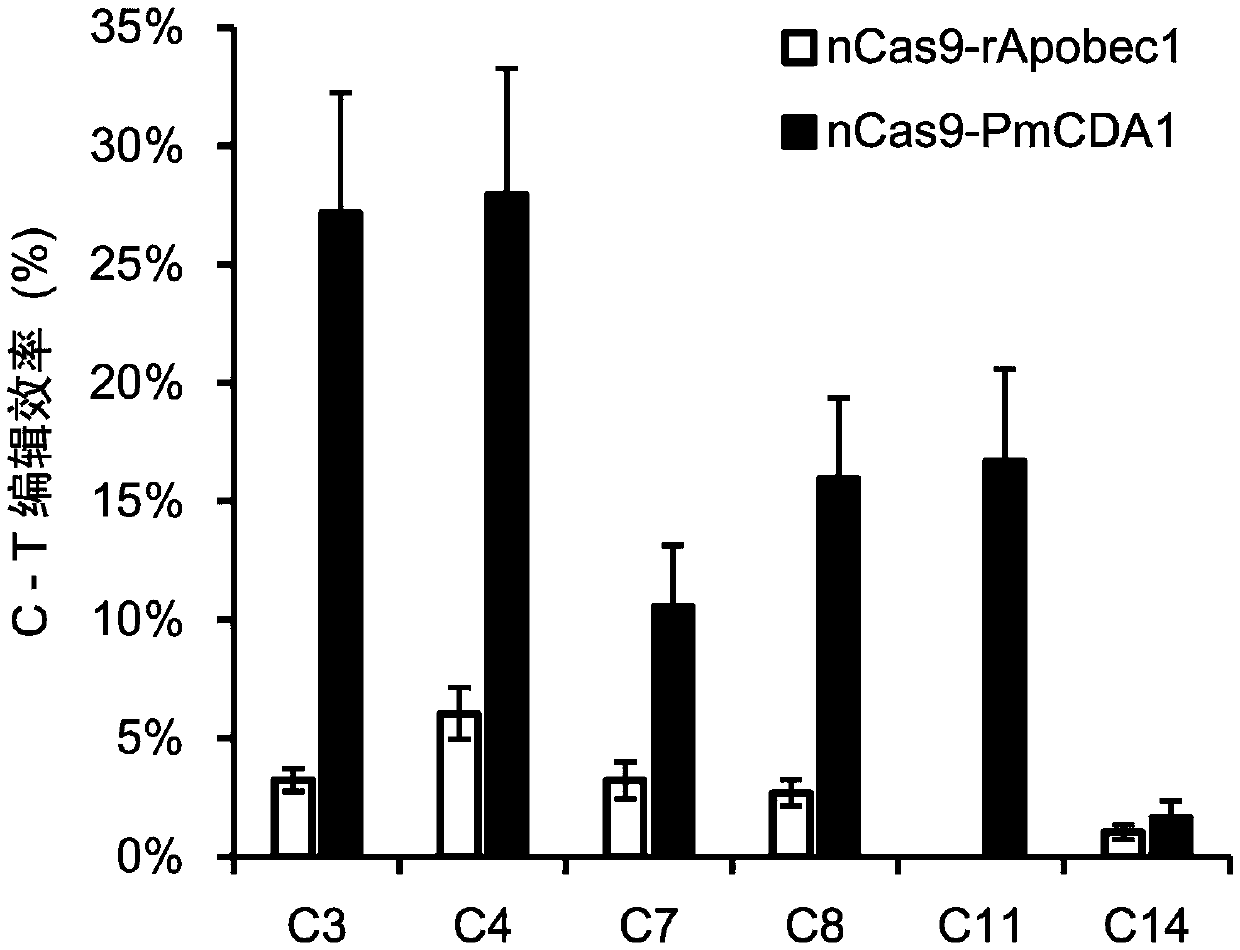
[0074] Example 2 High-throughput identification of rice endogenous gene cytosine-directed base editing efficiency based on STU nCas9-PmCDA1 system
[0075] 1. Rice endogenous gene sgRNA design and STU nCas9-PmCDA1 base editing recombinant expression vector construction
[0076] For rice OsCDC48 (LOC_Os03g05730) and OsROC5 (LOC_Os02g45250) coding genes, the 5'-NGG-3'PAM site was searched, and the 20bp sequence upstream of the PAM was selected to design sgRNA (Table 1).
[0077] Table 1 Design, synthesis and detection information of rice endogenous gene sgRNAcrRNA
[0078]
[0079] According to the designed sgRNA site nucleic acid sequence, artificially synthesize the corresponding forward and reverse oligonucleotide chains, the specific sequence is as follows (uppercase base sequence represents the designed site-specific guide sgRNA site; lowercase base sequence represents the site with Backbone vector complementary cohesive ends):
[0080] BE-OsCDC48-sgRNA01-F (Seq ID No....
Embodiment 3
[0095] Example 3 Based on the STU nCas9-PmCDA1 system, the creation and efficiency analysis of rice endogenous gene cytosine-directed base editing regenerated plants
[0096] 1. Agrobacterium transformation of rice endogenous gene STU nCas9-PmCDA1-sgRNA base editing recombinant expression vector
[0097]The STU nCas9-PmCDA1-OsCDC48-sgRNA01, STU nCas9-PmCDA1-OsROC5-gRNA04, and STU nCas9-PmCDA1-OsROC5-gRNA05 recombinant expression vectors successfully constructed in Example 2 and detected in rice protoplasts for directional modification activity were passed The competent cells of Agrobacterium EHA105 were transformed by the heat shock method, spread on LB solid medium containing 50 mg / L kanamycin and 50 mg / L rifampicin, and cultured in the dark at 28°C for 2 days to obtain positive clones. Positive clones were activated in LB liquid medium containing 50 mg / L kanamycin and 50 mg / L rifampicin for subsequent transformation.
[0098] 2. Agrobacterium-mediated rice callus transforma...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com