Halohydrin dehalogenase mutant and application thereof
A halohydrin dehalogenase and mutant technology, applied in the biological field, can solve the problems of excellent catalytic performance, inability to meet industrial production, and few industrial production.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0047] Example 2. Homology Modeling
[0048] Using the crystal structures of HheC from Agrobaterium radiobater AD1 and HheB from Corynebacteriumsp.N-1074 as templates, Modeller 9.18 was used for HHDH His The three-dimensional structure was homologously modeled, and then CHARMM27 was used to optimize the model. The evaluation program ProCheck was used to evaluate the model to evaluate the reliability of the obtained model. The modeling results were analyzed using the software PyMOL 1.2rl. Ended up with high quality HHDH His structural model. HHDH His The evaluation results of the model showed that: 89.8% were located in the optimal region, 8.2% were located in other allowed regions, 2.0% were located in the general allowed region, and no residues were located in the disallowed region, which indicated that the model was reliable. Through sequence alignment and structural analysis, it was found that HHDH His The catalytic triad structure is Ser116-Tyr129-Arg133.
Embodiment 3
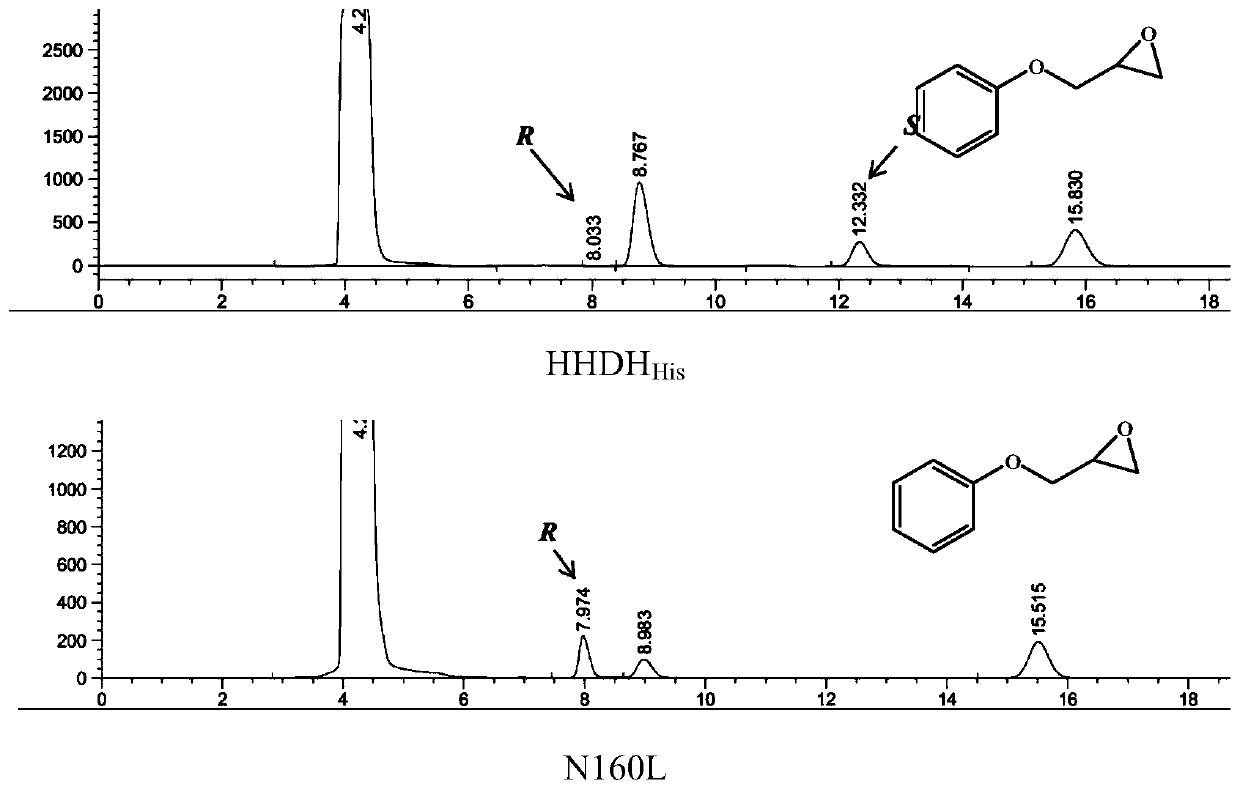
[0049] Example 3. Molecular docking
[0050] Using the molecular docking software Autodock 4.0, the halohydrin dehalogenase HHDH His Molecular docking to the active center of racemic PGE; select distance from substrate molecule Nearby sites were subjected to site-directed saturation mutagenesis, and the final selected mutation sites were Gln159, Asn160, Phe161, Tyr167, Phe168, and Pro169. The primers used for site-directed mutagenesis are shown in Table 3.
[0051] Table 3 PCR primers used for mutation
[0052]
[0053] N stands for A, T, G or C
[0054] Site-directed saturation mutation was performed on the selected 6 sites, and clones were selected for activity screening at each site. First, false positive strains with no activity were eliminated through high-throughput screening, and then mutant strains with improved stereoselectivity were screened through liquid phase detection. by HHDH His Screening of the 10 saturation mutant library, at the Phe161, Tyr167, and P...
Embodiment 4
[0055] Example 4. Induced expression of halohydrin dehalogenase and mutants thereof
[0056] HHDH His and its mutants N160L and Q159L were induced to express. Cultivate the recombinant halohydrin dehalogenase engineered bacteria at 37°C and 200r / min for 10h, add the Escherichia coli with the recombinant expression plasmid to 50mL LB liquid medium at an inoculum size of 1%, and add a final concentration of 50mg / min. L of kanamycin, cultured to OD at 37°C and 200r / min 600 If the value is 0.6-0.8, IPTG inducer is added to make the final concentration 0.15mM to induce its protein expression. Induce the culture at 28°C and 200r / min for about 10h, centrifuge at 5000×g for 5min, collect the cells, resuspend the bacteria with pH 8.0 phosphate buffer (200mM), and ultrasonically disrupt the cells.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com