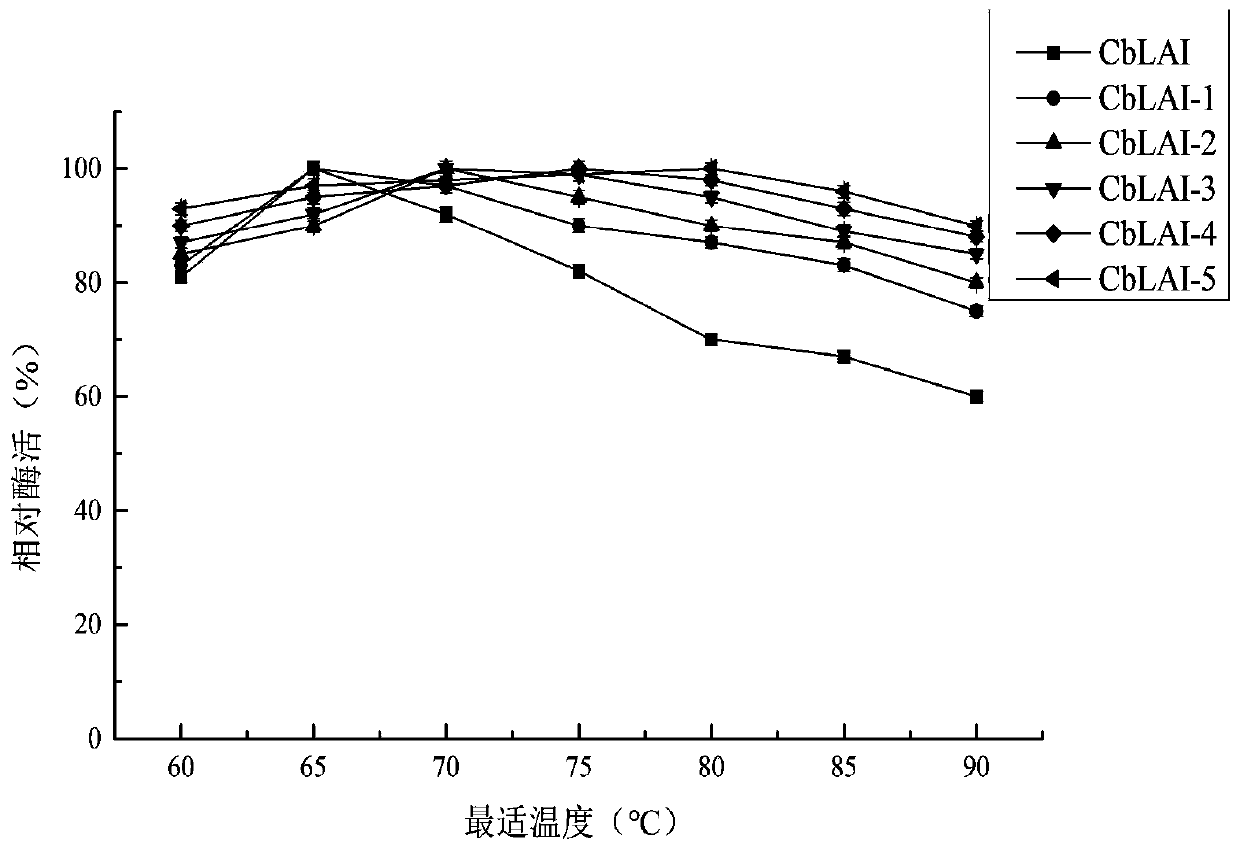
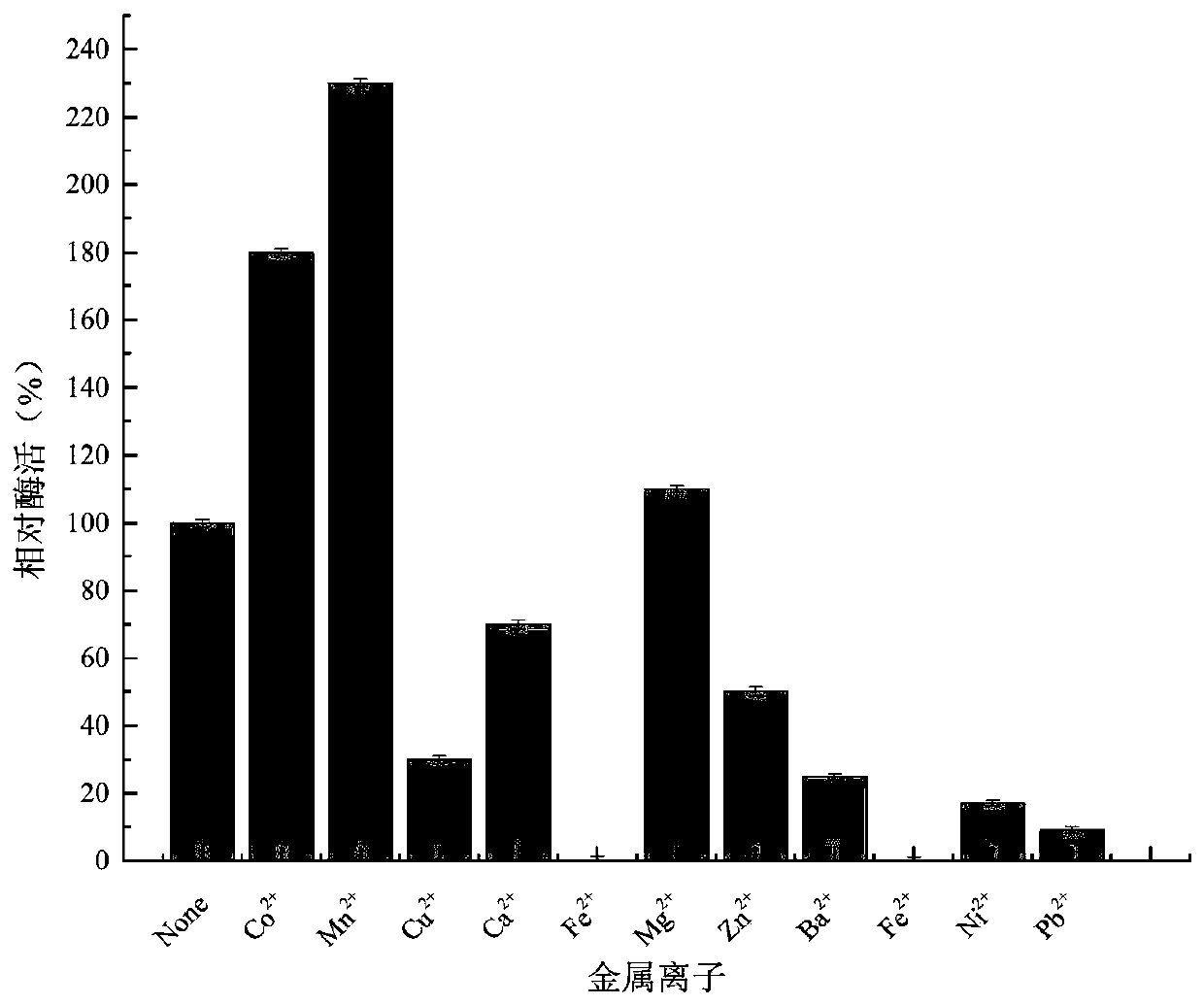
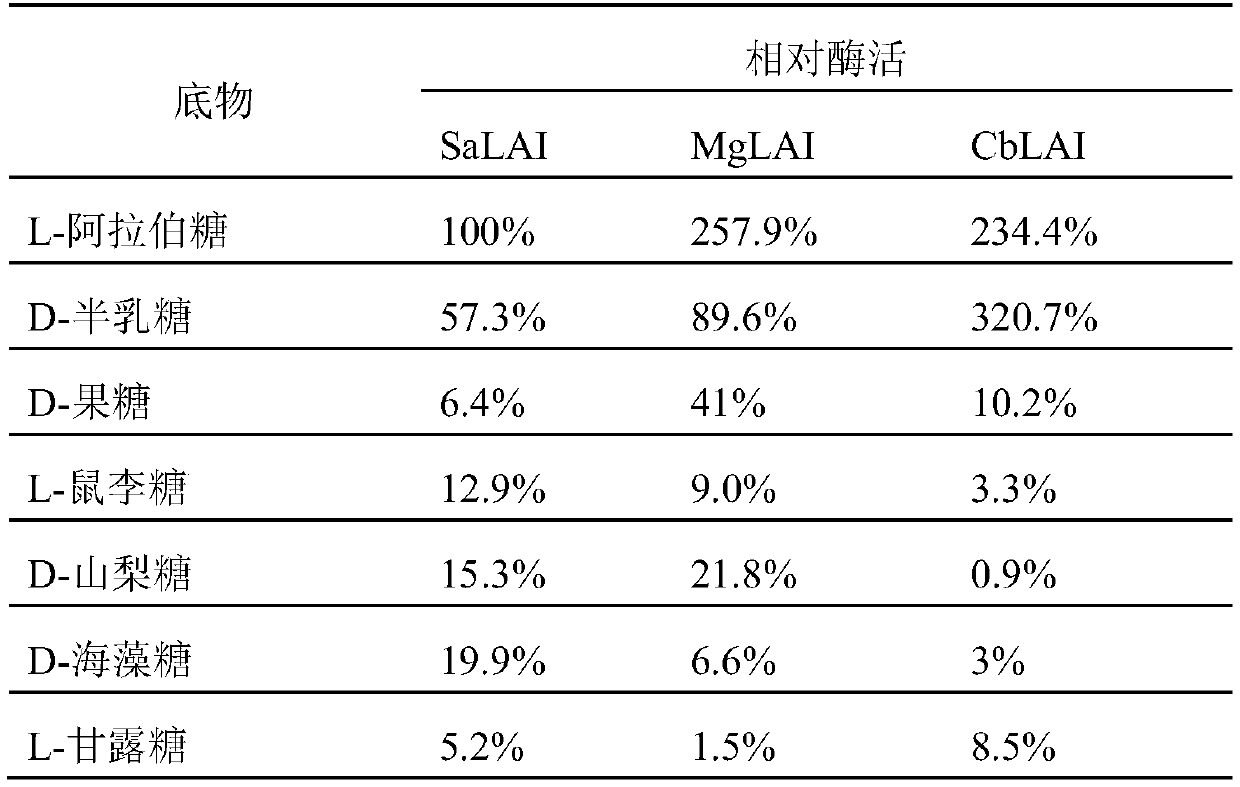
L-arabinose epimerase mutant and application thereof
A technology of epimerase and arabinose, which is applied in the field of L-arabinose epimerase mutants, can solve the problems of catalytic efficiency to be improved, low substrate concentration, unfavorable industrial production, etc., and achieve good industrial application foreground effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Embodiment 1: the screening of novel enzyme
[0024] 1. Source of enzyme and construction of recombinant bacteria
[0025] The sugar isomerases obtained from the NCBI database were derived from Salegentibacteragarivorans (GenBank No. WP_093303409.1), Massilia glaciei (GenBank No. WP_106758181.1), Chitinophagaceae bacterium (GenBank No. WP_153799928.1), and named SaLAI, MgLAI and CbLAI. According to the amino acid sequence, the codon was optimized according to the codon preference of Escherichia coli, and three selected nucleotide sequences were synthesized by the method of total synthesis through the conventional operation of genetic engineering, such as SEQ ID NO.2, SEQ ID NO.4 and shown in SEQ ID NO.6; the amino acid sequences encoding the enzyme are shown in SEQ ID NO.1, SEQ ID NO.3 and SEQ ID NO.5 respectively. Add 6×His-tag tags at the end of the nucleic acid sequence, add restriction sites XbaI and XhoI at both ends, clone the gene into the XbaI and XhoI sites c...
Embodiment 2
[0039] Example 2: Construction and screening of CbLAI single point mutants
[0040] 1. Mutant construction
[0041] According to the CbLAI parental sequence (the amino acid sequence is shown in SEQ ID NO.5, and the nucleotide sequence is shown in SEQ ID NO.6), the mutation primers for site-directed mutation were designed, and the recombinant vector pET28b / CbLAI was used as a template by using rapid PCR technology. To introduce a single mutation at position 97, the primers are:
[0042] Forward primer 97G: CCGGCAAAGATGTGGATCGGT NNN CTAAAAGTAC (the underline is the mutated base)
[0043] Reverse primer 97G: GTACTTTTAG NNN ACCGATCCACATCTTTGCCGG (the underline is the mutated base)
[0044] PCR reaction system: 2×Phanta Max Buffer (containing Mg 2+ ) 25 μL, dNTPs 10 mM, forward primer 97G 2 μL (5 pmol / μL, the same below), reverse primer 97G 2 μL (5 pmol / μL, the same below), template DNA 1 μL (20ng / μL, the same below), Phanta Max Super- Fidelity DNA Polymerase 50U with ddH a...
Embodiment 3
[0056] Example 3: Construction and screening of CbLAI double-site mutants
[0057] According to the sequence of the single mutant CbLAI-1 constructed in Example 2, the mutation primers for site-directed mutation were designed. Using the rapid PCR technology, the recombinant vector pET28b / CbLAI-1 was used as a template to introduce a single mutation at position 189. The primers were:
[0058] Forward primer 189N: TTCGGCGAC NNN ATGAGACAAGTAGCAGTGACAG (the underline is the mutated base)
[0059] Reverse primer 189N: CTGTCACTGCTACTTGTCTCAT NNN GTCGCCGAA (the underline is the mutated base)
[0060] PCR reaction system: 2×Phanta Max Buffer (containing Mg 2+ ) 25μL, dNTPs 10mM, forward primer 189N 2μL, reverse primer 189N 2μL, template DNA 1μL, Phanta Max Super-Fidelity DNA Polymerase 50U, add ddH 2 0 to 50 μL.
[0061] The PCR amplification conditions were 95°C for 3min; (95°C for 15s, 62.5°C for 15s, 72°C for 6.5min) for 30 cycles; 72°C for 5min.
[0062] The PCR product wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com