Cell screening model for label-free histamine receptor H2
A histamine receptor, label-free technology, applied in the field of cell screening, can solve the problems of cumbersome operation, low throughput, long experimental period, etc., and achieve the effects of pathway integration research and simple operation, high temporal and spatial resolution, and short experimental period.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
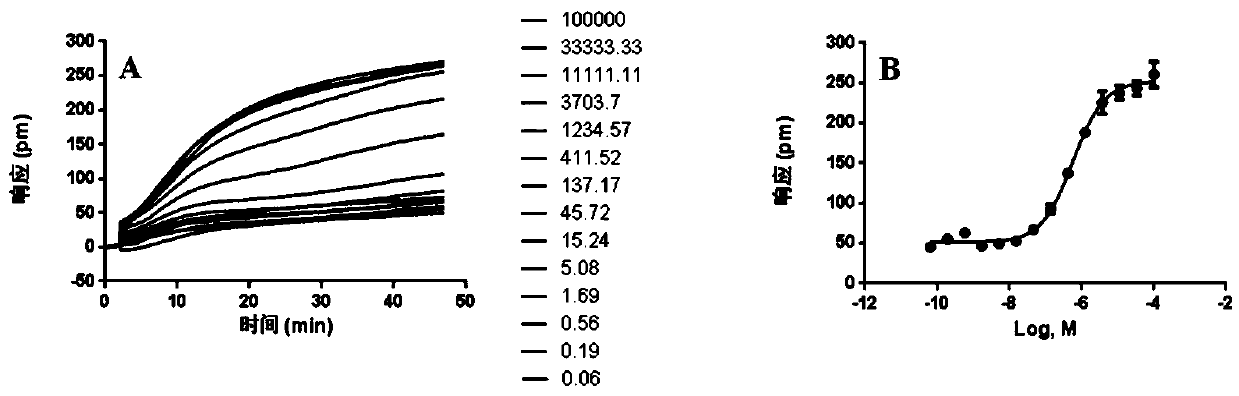
[0031] Example 1: DMR characteristic signal spectrum of agonist histamine on HEK-293-H2 cells
[0032] Human embryonic kidney cells HEK-293-H2 cells were derived from the cell bank independently constructed by the laboratory, the inverted microscope was purchased from OLYMPUS, histamine and cimetidine were purchased from Bailingwei and AMQUAR respectively. The cell culture plate is an Epic optical biosensing 384 microwell plate, purchased from Corning, and the detection platform is the third generation of Corning Imager, the detected signal is the wavelength shift caused by cell dynamic mass reset (DMR).
[0033] HEK-293-H2 cells in the logarithmic growth phase were seeded in a cell-compatible 384 microwell plate, the medium used was DMEM (#SH30022.01B, Thermo), the inoculation volume of each well was 40 μL, and each well was inoculated The number of cells is 2.0 x 10 4 Firstly, place the inoculated cell plate in a cell incubator and culture it for 20-22 hours until the cel...
Embodiment 2
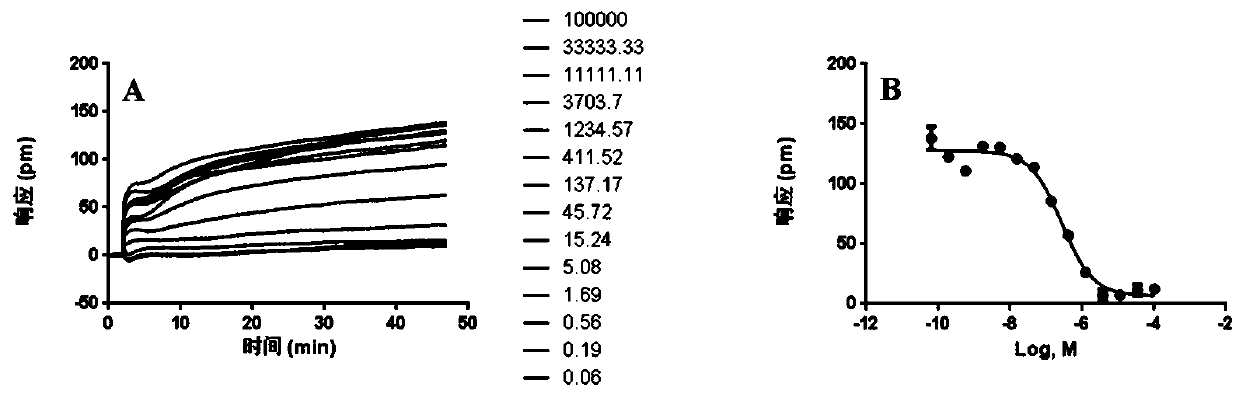
[0034] Example 2: DMR characteristic signal spectrum of antagonist cimetidine on HEK-293-H2 cells
[0035] HEK-293-H2 cells in the logarithmic growth phase were seeded in a cell-compatible 384 microwell plate, the medium used was DMEM (#SH30022.01B, Thermo), the inoculation volume of each well was 40 μL, and each well was inoculated The number of cells is 2.0 x 10 4 Firstly, place the inoculated cell plate in a cell incubator and culture it for 20-22 hours until the cell confluency reaches about 95%, and conduct an activity test. Replace the cell culture solution in the microwell plate with Hank's balanced salt solution (containing 20mM HEPES), and add a volume of 30 μL to each well. After adding, place in Equilibrate on the imager for 1 hour; re-scan the baseline for 2 minutes, add different concentrations of cimetidine into the microwell plate, the volume of each well is 10 μL, the concentration is 100000nM, 33333.33nM, 11111.11nM, 3703.7nM, 1234.57nM, 411.52 nM, 137.17nM...
Embodiment 3
[0036] Example 3: Desensitized DMR characteristic signal spectrum of HEK-293-H2 cells
[0037] HEK-293-H2 cells in the logarithmic growth phase were seeded in a cell-compatible 384 microwell plate, the medium used was DMEM (#SH30022.01B, Thermo), the inoculation volume of each well was 40 μL, and each well was inoculated The number of cells is 2.0 x 10 4 Firstly, place the inoculated cell plate in a cell incubator and culture it for 20-22 hours until the cell confluency reaches about 95%, and conduct an activity test. Replace the cell culture solution in the microwell plate with Hank's balanced salt solution (containing 20mM HEPES), and add a volume of 30 μL to each well. After adding, place in Equilibrate on the imager for 1h; add different concentrations of histamine into the microwell plate to pretreat HEK-293-H2 cells for 1h, the volume of each well is 10μL, and the concentration is 100000nM, 33333.33nM, 11111.11nM, 3703.7nM, 1234.57nM, 411.52nM, 137.17nM, 45.72nM, 15.2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com