Genetically engineered bacterium and application thereof to preparation of 9alpha,22-dihydroxy-23,24-bisnorcholest-4-ene-3-ketone
A technology of genetically engineered bacteria and steroids, applied in the field of genetic engineering, can solve the problems of difficulty in separation and purification and high production costs, and achieve the effects of reducing pollution, improving production efficiency and product quality, and reducing production costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Embodiment 1: Construction of gene inactivation strain
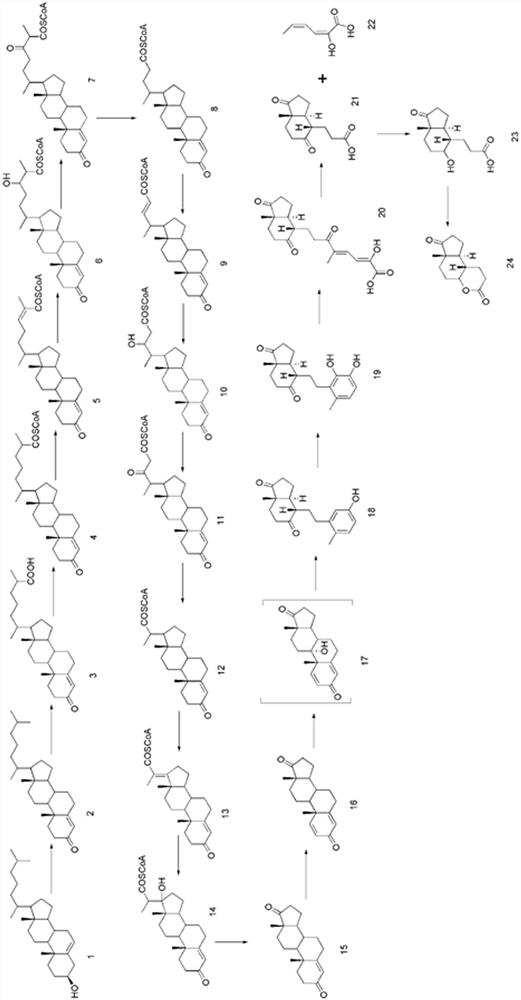
[0022] 3-sterone-∆ 1 -Dehydrogenase encoding gene ( kstd1, kstd2, kstd3 ) as shown in SEQ ID NO: 1, SEQ ID NO: 2, SEQ ID NO: 3, to start the strain Mycobacteriu. smegmatis mc 2 The genome of 155 (strain number: ATCC 700084) was used as a template for PCR amplification, and the inactivation kstd1, kstd2 and kstd3 strains.
[0023] This study uses CRISPR-Cas12a-assisted recombineering (references: Yan MY, Yan HQ, Ren GX, Zhao JP, Guo XP, Sun YC. CRISPR-Cas12a-assisted recombineering in bacteria[J]. Appl Environ Microbiol , 2017, 83(17): e00947-17) to construct a knockout strain. first in kstd1 The 31-base sequence targeting the lagging strand was selected as the gene-specific crRNA, and the pCR-Hyg plasmid was used as a template to amplify with two pairs of primers, KstD1-F / R and pCR-Hyg-F / R, to obtain two The plasmid fragments were ligated and transformed and sent for sequencing, and the correct extract...
Embodiment 2
[0053] Embodiment 2: Construction of gene expression strain
[0054] 1. Expression plasmid construction
[0055] Primers cxgAB-F and cxgAB-R were used to amplify from Mycobacterium Acetyl-CoA acetyltransferase / thiolase gene (cxgA) and DNA binding protein gene (cxgB) in the sp. NRRL B-3805 genome, after obtaining the cxgAB fragment with a 15bp homology arm with the plasmid pMV261, and expressing Plasmid pMV261 was digested and purified by EcoRI and HindIII to obtain a single-fragment connection, and the recombinant expression plasmid 261-cxgAB was obtained.
[0056] PCR system:
[0057] 5×Phusion GC Buffer 10 μl
[0058] 2mM dNTPs 5 μl
[0059] Primer F 1 μl
[0060] Primer R 1 μl
[0061] Template DNA 50-100ng
[0062] DMSO 1.5 μl
[0063] Phusion 0.5 μl
[0064] wxya 2 O to 50 μl
[0065] PCR program: 3 min at 98°C; denaturation at 98°C for 10 s, annealing at 58°C for 20 s, extension at 72°C for 30 s, 30 cycles; 10 min at 72°C. The primer sequences used are:
...
Embodiment 3
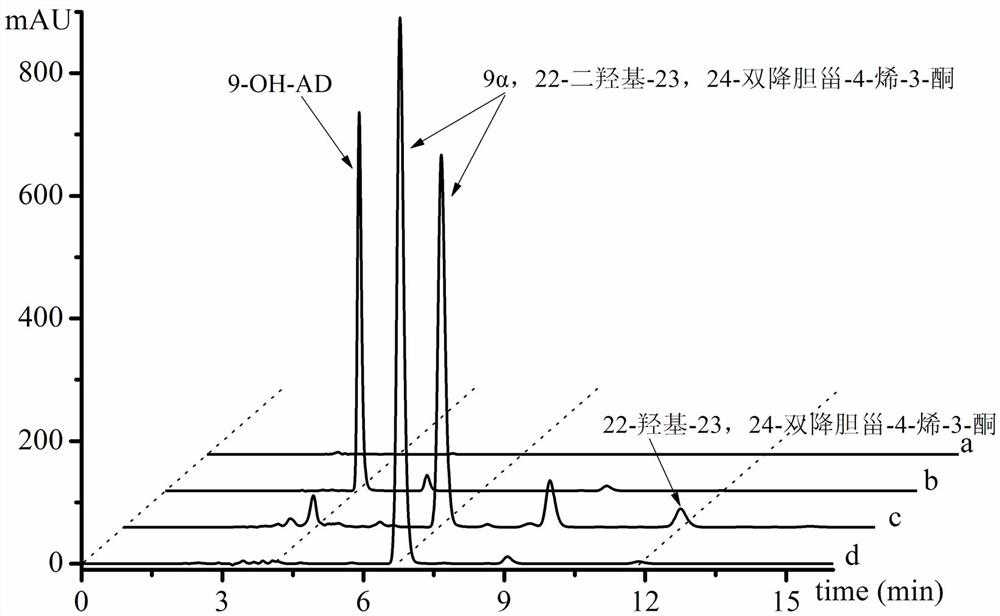
[0071] Example 3: Fermentative production of 9α, 22-dihydroxy-23, 24-bisnorcholest-4-en-3-one
[0072]Seed medium: glucose 6 g / L, yeast powder 15 g / L, NaNO 3 5.4 g / L, glycerol 2 g / L, NH 4 h 2 PO 4 0.6g / L, kanamycin 0.1 mg / L, pH 7.5, sterilized at 115°C for 30 minutes.
[0073] Fermentation medium: defatted soybean powder 10 g / L, corn steep liquor 5 g / l, diammonium hydrogen phosphate 3 g / L, soybean oil 150 g / L, kanamycin 0.1 mg / L, pH 7.5, 121°C Sterilize for 30 minutes.
[0074] Fermentation culture steps:
[0075] 1. Cultivate LB plates (tryptone: 10g / L, yeast extract: 5g / L, sodium chloride: 10g / L, agar: 15 g / L) at 37°C for 72 hours to activate the strains;
[0076] 2. Inoculate from the activated plate into the seed medium, cultivate at 180 rpm and 37°C for 3 days; sample the seed liquid under aseptic conditions for sampling and microscopic examination, and inoculate without any bacteria in the microscopic examination;
[0077] 3. Inoculate 10% of the inoculum into 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com