Adenosine engineering bacterium as well as construction method and application thereof
A technology of adenosine engineering and construction method, which is applied in the fields of application, genetic engineering, botanical equipment and methods, etc., and can solve the problems of difficult genetic engineering transformation of Bacillus subtilis, increased fermentation cost, bacterial contamination and reverse irrigation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
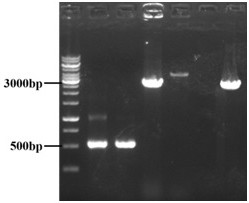
[0069] Integration of the purine nucleoside operon purEKBCSQLFMNHD into Escherichia coli
[0070] 1. Use the premier5 software to design the upstream homology arm primers (UP-yjiV- S, UP-yjiV-A) and downstream homology arm primers (DN-yjiV-S, DN-yjiV-A) and PCR amplification of the upstream and downstream homology arm fragments. Then with B. subtilis The XGL genome was used as a template to design the upstream and downstream primers (purEKB-S, purEKB-A) of the target gene purEKB, and the target fragment was amplified by PCR. The promoter is designed in the downstream primer of the upstream homology arm and the upstream primer of the target fragment, and the recognition target sequence is designed in the downstream primer of the target fragment and the upstream primer of the downstream homology arm (see Table 1 for the above-mentioned primers). The length of the upstream homology arm involved above is 477bp, the downstream homology arm is 499bp, and the length of the target ...
Embodiment 2
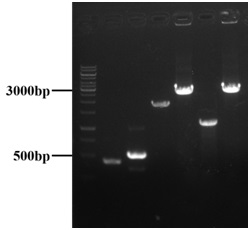
[0080] The yfkN gene was linked to the strong promoter Ptrc and integrated into the pseudogene mbhA. The difference from the above-mentioned genes is that the yfkN gene sequence is relatively large, and if the one-time integration efficiency is not high, segmental integration is required. Therefore, the yfkN gene is divided into two segments, yfkN-UP and yfkN-DN. The specific steps are divided into the following steps:
[0081] 1. Use the Premier5 software to design the upstream homology arm primers (UP-mbhA-S, UP-mbhA-A) and downstream homology arm primers (DN-mbhA- S1, DN-mbhA-A) and PCR amplification of its upstream and downstream homology arm fragments. Then with Bacillus subtilis The XGL genome was used as a template to design the upstream and downstream primers (yfkN-UP-S, yfkN-UP-A) of the target gene yfkN-UP, and the target fragment was amplified by PCR. The promoter is designed in the downstream primer of the upstream homology arm and the upstream primer of the ...
Embodiment 3
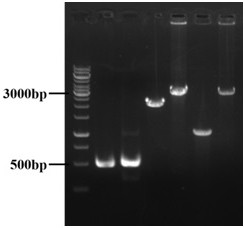
[0090] Knockout of genes guaB, add, amn, ygdH and deoD related to adenosine precursor and adenosine degradation pathway
[0091] Knockout of guaB gene
[0092] 1. Using Escherichia coli E.coli W3110 as a template, design upstream homology arm primers ((UP-guaB-S, UP-guaB-A) and downstream homology arm primers (DN-guaB- S, DN-guaB-A), the upper and lower homology arms were amplified by PCR, and the knockout fragment of the guaB gene (upstream homology arm-downstream homology arm) was obtained by fusion with PCR overlapping technology. For specific primer sequences, see Table 1 below.
[0093] 2. Construction of pGRB-guaB, the DNA fragments gRNA-guaB-S and gRNA-guaB-A of the target sequence used in the process of constructing pGRB-guaB are shown in Table 1 below.
[0094] 3. Use the last strain that lost pGRB to prepare competent cells. For the preparation method of competent cells, refer to the above. Electroporation was performed, and the knockout fragment of the guaB gene ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com