T7-RNA polymerase mutant and application thereof
A technology of T7-RNA, 1.T7-RNA, applied in the field of nucleic acid tool enzymes and nucleic acid biology, can solve the problem of maintaining high-efficiency transcription RNA product heterogeneity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
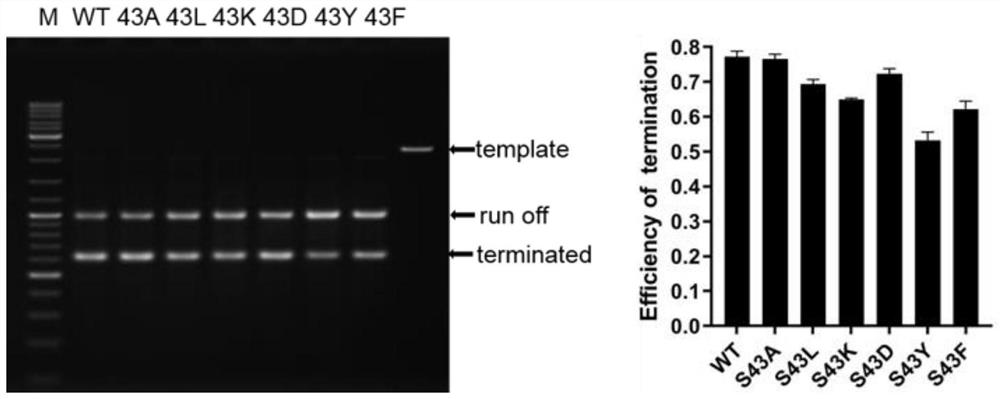
[0025] Example 1: In vitro termination efficiency detection using T7--RNA polymerase mutant
[0026] (1) Expression and purification of T7-RNA polymerase mutant
[0027]The T7-RNA polymerase mutants S43A, S43L, S43K, S43D, S43Y and S43F were respectively constructed by molecular cloning, and then the prokaryotic expression vector pQE82L containing these mutants was transformed into the E.coli BL21 expression strain, and the bacteria were picked and expanded for cultivation , place the bacteria in LB medium containing 100 μg / ml ampicillin and culture on a shaker at 37°C until the OD600 value is close to 1.2, and then add isopropyl-β-D-thiogalactopyranoside at a final concentration of 0.5mM (IPTG) was induced to express on a shaking table at 30°C for 4h, and then centrifuged at 4°C and 5000rpm for 20min to collect the cell pellet, and then the cell was fully resuspended in a solution containing 300mM NaCl, 20mM Tris-HCl (pH=7.5), 0.5mg / ml lysozyme, 0.5mM DTT lysate, frozen at ...
Embodiment 2
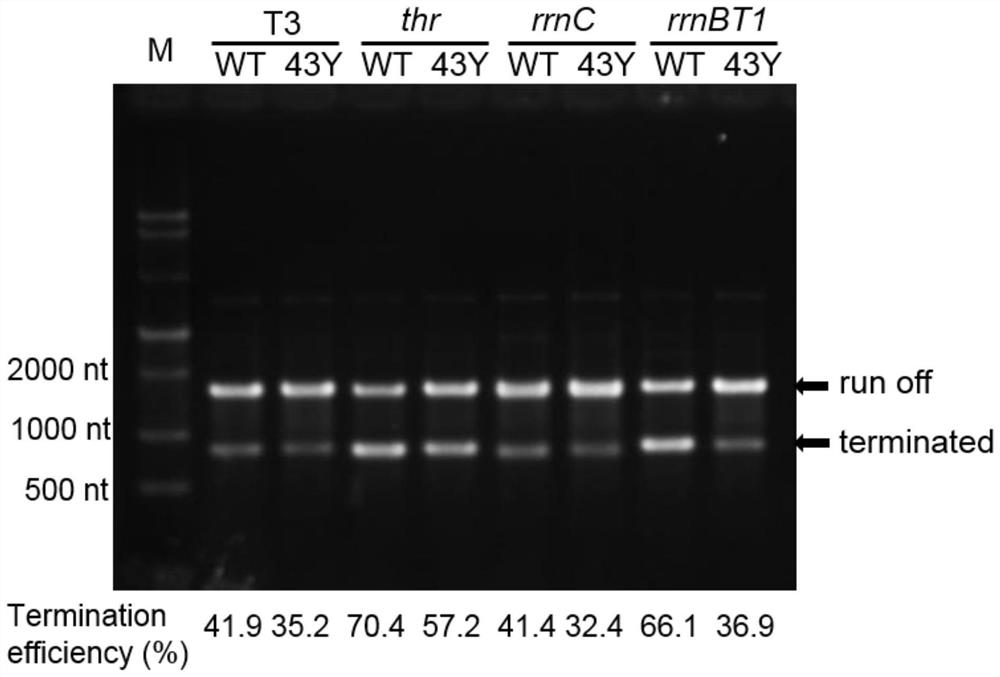
[0038] Example 2: Comparison of T7-RNA polymerase mutant S43Y and wild type for class I termination efficiency from different sources
[0039] (1) Acquisition of transcription reaction template
[0040] The sequences of several class I terminators T3 TΦ, thr attenuator, rrnBT1 terminator and rrnC terminator reported in T3 phage and Escherichia coli by molecular cloning (respectively see the sequence table SEQ ID NO.3-6 ) to replace the neck loop structure sequence of the class I terminator T7 TΦ in the vector pETsnrs-1 (see the sequence listing SEQ ID NO.7). Carry out PCR amplification with the general primer of embodiment 1 after the successful construction of vector, then utilize DNA purification kit Clean&Concentrator TM -5 Purify and measure the concentration of the PCR product.
[0041] (2) Detection of transcription termination efficiency in vitro
[0042] The in vitro transcription reaction (10μL) contained 40mM Tris-HCl (pH=8.0), 16mM MgCl 2 , 2mM spermidine, 5mM D...
Embodiment 3
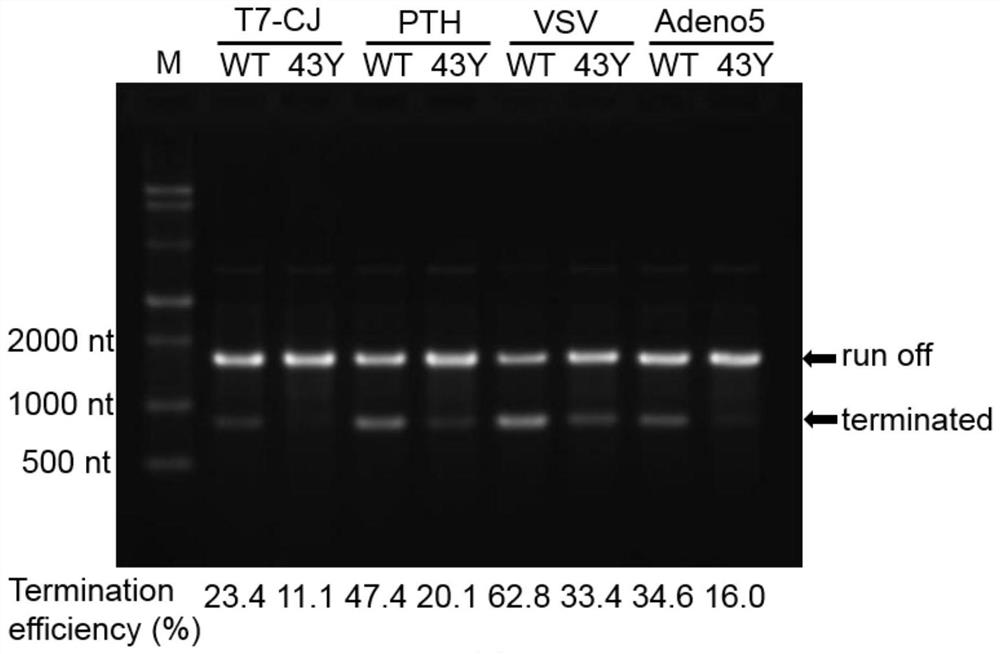
[0043] Example 3: Comparison of T7-RNA polymerase mutant S43Y and wild type on the termination efficiency of class II from different sources
[0044] (1) Acquisition and purification of transcription reaction templates
[0045] By means of molecular cloning, the class II termination signal sequences (respectively see SEQ ID NO. There is a termination sequence. Similarly, the constructed vector was amplified by PCR using the universal primers described in Example 1, and then the PCR product was purified and its concentration was determined using a DNA purification kit.
[0046] (2) Detection of transcription termination efficiency in vitro
[0047] The in vitro transcription reaction components contained 40mM Tris-HCl (pH=8.0), 16mM MgCl 2 , 2mM spermidine, 5mMDTT, 4mM ATP, GTP, CTP, UTP, 0.3μL RNase inhibitor, 0.2μL pyrophosphatase, 50nM RNA polymerase, 14nM PCR template, add DEPC water to 10μL. The transcription reaction conditions were incubation at 37°C for 1 h, followe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com