Sumoylation peptide fragment enrichment method based on desumoylation enzyme and SAX
A peptide and pre-enrichment technology, which is applied in chemical instruments and methods, peptide preparation methods, peptides, etc., can solve problems such as inability to accurately reflect the real modification state of proteins, and achieve improved identification coverage and high enrichment selectivity , reduce the effect of identifying false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
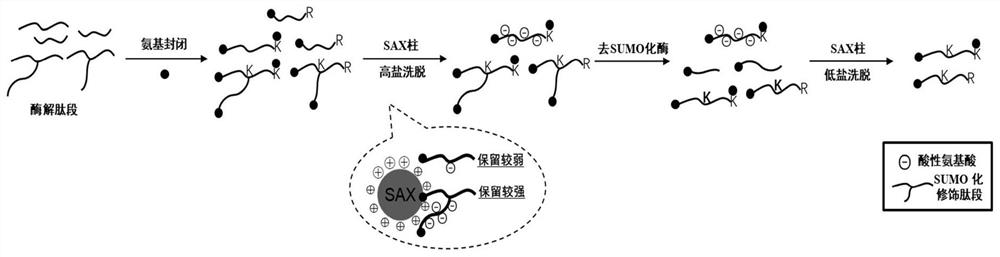
[0037] Such as figure 1 As shown, first digest the basic site of the protein, and then block the N-terminal of the peptide and the free amino group of the side chain at the peptide level. The retention in anion-exchange chromatography is stronger than that of non-SUMOylated peptides, so anion-exchange chromatography is used to pre-enrich the SUMOylated peptides with strong retention, and then the collected SUMOylated peptides are deSUMOlated. After de-SUMOylation, the retention of de-SUMOylated peptides in anion-exchange chromatography becomes weaker, so that the enrichment of SUMOylated peptides is achieved in the second anion-exchange chromatography.
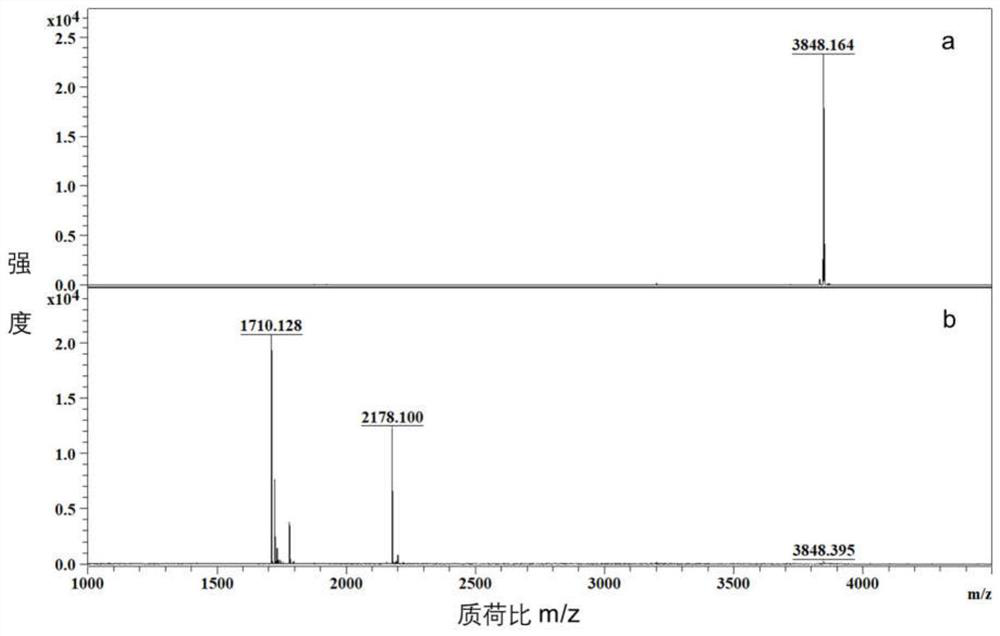
[0038] The SUMO1 standard peptide with the sequence ELGMEEEDVIEVYQEQTGG(19)-LLVHMGLLKSEDKVK(9) was used as the sample, dissolved in 50mM ammonium bicarbonate, and digested with deSUMOlase SENP1, wherein the amount of enzyme was 1 / 20 of the sample mass, and the temperature was After enzymatic hydrolysis for 12 hours at 37°C, p...
Embodiment 2
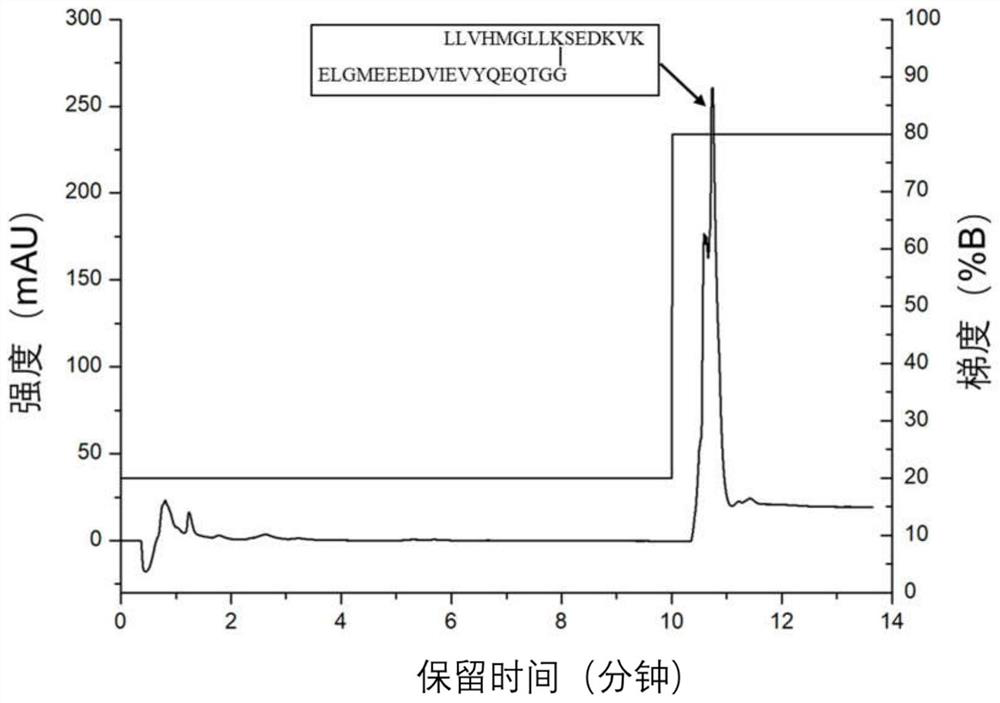
[0040]Redissolve 10 μg of SUMO1 standard peptides in phase A, load the sample onto a tertiary amine-based ion exchange column (4.6mm i.d×5cm) for isocratic elution, elution gradient (V / V): 0-10min, 20% B; 10-20min, 80% B, flow rate is 1.0mL / min. Phase A: 20% acetonitrile + 1 mM pH 8 Tris buffer; Phase B: 20% acetonitrile + 1 mM pH 8 Tris buffer + 500 mM sodium chloride. Such as image 3 As shown, after the unretained fraction 10 minutes before elution, the fraction containing the SUMO1-labeled peptide 10 minutes later was collected, desalted, and lyophilized. It was redissolved in 0.1% formic acid and analyzed by mass spectrometry. It can be seen that the SUMO1 standard peptide was effectively retained and enriched in anion exchange chromatography.
Embodiment 3
[0042] 10 μg of SUMO1ylated standard peptides were digested with deSUMOlase, loaded onto a tertiary amine-based ion exchange column (4.6mm i.d×5cm) for isocratic elution, elution gradient: 0-10min, 20% B; 10-20min, 80% B, flow rate is 1.0mL / min. Phase A: 20% acetonitrile + 1 mM pH 8 Tris buffer; Phase B: 20% acetonitrile + 1 mM pH 8 Tris buffer + 500 mM sodium chloride. Such as Figure 4 As indicated, the non-retained fractions in the first 10 minutes and the retained fractions after 10 minutes were collected, desalted, and freeze-dried. Redissolved in 0.1% formic acid and analyzed by mass spectrometry, the de-SUMOylated peptide LLVHMGLLKSEDKVK after being digested by de-SUMOlase does not retain the outflow in anion exchange, the original SUMO1-ylated standard peptide and the peptide obtained after digestion ELGMEEEDVIEVYQEQTGG was still retained in anion exchange, indicating that the effective enrichment and identification analysis of SUMOylated peptides was successfully ac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com