Crassostrea gigas DM9 structural domain-containing recombinant protein rCgDM9CP-6, and preparation method and application thereof
A technology of recombinant protein, long oyster, applied in the field of molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
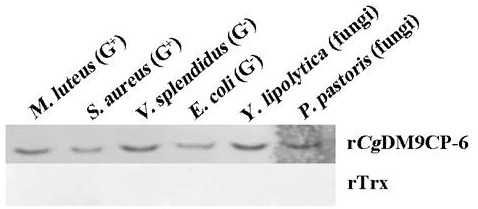
[0036] Experimental example 1: Detection of the binding activity of the recombinant protein rCgDM9CP-6 containing the DM9 domain of the oyster of the present invention to bacteria
[0037] Based on the western blotting method, the recombinant protein rCgDM9CP-6 was detected against two Gram-negative bacteria (Vibrio brilliant and Escherichia coli), two Gram-positive bacteria (Micrococcus luteus and Staphylococcus aureus) and two fungi. Saccharomyces rovia and Pichia pastoris binding activity. The sources of the strains used are as follows: Vibrio splendidus ( Vibrio splendidus JZ6) was purchased from Beijing Microorganism Culture Collection Center, Escherichia coli ( Escherichia coli ) was purchased from Beijing Quanshijin Company, Staphylococcus aureus ( Staphylococcus aureus ) was purchased from Beijing Microorganism Culture Collection Center, Micrococcus luteus ( Micrococcus luteus ) was purchased from Beijing Microorganism Culture Collection Center, Yarrowia sacchar...
experiment example 2
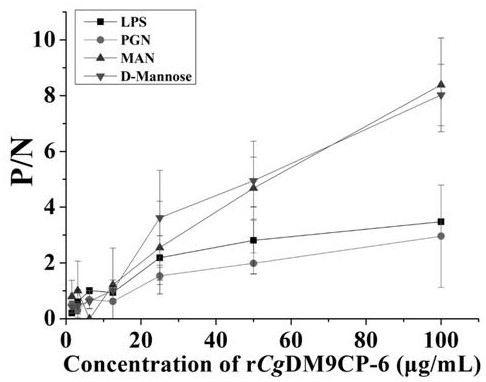
[0057] Experimental Example 2: Detection of sugar-binding activity of the recombinant protein rCgDM9CP-6 containing the DM9 domain of oyster of the present invention
[0058] The enzyme-linked immunosorbent assay (ELISA) test was used to detect the combination of the recombinant protein rCgDM9CP-6 containing the DM9 domain of the present invention with various PAMPs. The four sugars used, D-Mannose, Mannose, LPS and PGN, were purchased from Sigma.
[0059] The specific operation steps are as follows:
[0060] (1) Na 2 CO 3 with NaHCO 3 Prepare the coating solution with a pH value of 7.6 according to the concentration of 15 mmol / L and 35 mmol / L, and dissolve the four sugars D-Mannose, Mannose, LPS and PGN to adjust the concentration to 125 μg / mL, and then add 100 μL, refrigerate overnight at 4°C;
[0061] (2) Discard the coating liquid and wash with TBS-T 4 times, 4 min each time;
[0062] (3) After washing, add 250 μL of 3% BSA to the wells and block them in a constant t...
experiment example 3
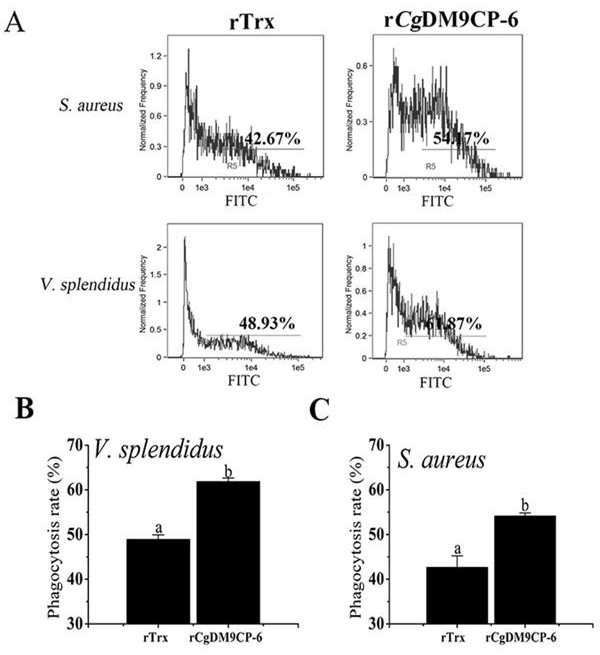
[0072] Experimental example 3: detection of phagocytosis-promoting activity of recombinant protein rCgDM9CP-6 containing DM9 domain in long oyster of the present invention
[0073] Two microorganisms (Vibrio splendidus and Staphylococcus aureus) were labeled with fluorescein isothiocyanate isomer (FITC), and then the phagocytic efficiency of oyster hemolymphocytes was detected by flow cytometry.
[0074] The strain source is as above.
[0075] The specific operation is as follows:
[0076] (1) Cultivate the above two microorganisms separately and collect the bacteria;
[0077] (2) Use formaldehyde to mix with microorganisms and fix for 10 min;
[0078] (3) Collect bacteria by centrifugation at 4,000 rpm for 10 min; 0.1 M NaHCO 3 After washing three times, incubate in 0.1 M NaHCO containing 0.1 mg / mLFITC 3 Incubate at 25°C for 2 h with gentle shaking;
[0079] (4) After centrifuging and discarding the supernatant, use TBS buffer to wash the microorganisms until they are co...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com