Detection method and kit for novel coronavirus (SARS-CoV-2)
A detection method and reagent technology, applied in the biological field, can solve problems such as time-consuming, time-consuming, labor-intensive, costly, unsafe, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0195] 1.1 SENA analysis targeting SARS-CoV-2 ORF1ab
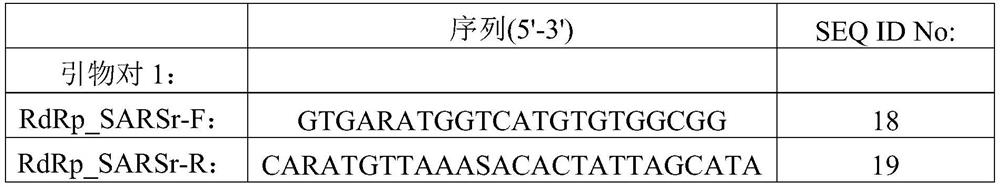
[0196] In this example, in order to screen a suitable guide RNA to target the sequence specifically amplified in SARS-CoV-2 ORF1ab, according to the primers recommended by China CDC (see primer pair 2, SEQ ID No: 20 and 21) and supporting The qRT-PCR amplified Taqman fluorescent probe sequence (5'-FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3', SEQ ID No: 8) determined the qRT-PCR amplification interval of ORF1ab.
[0197] The following crRNA sequences were designed from the amplified sequences:
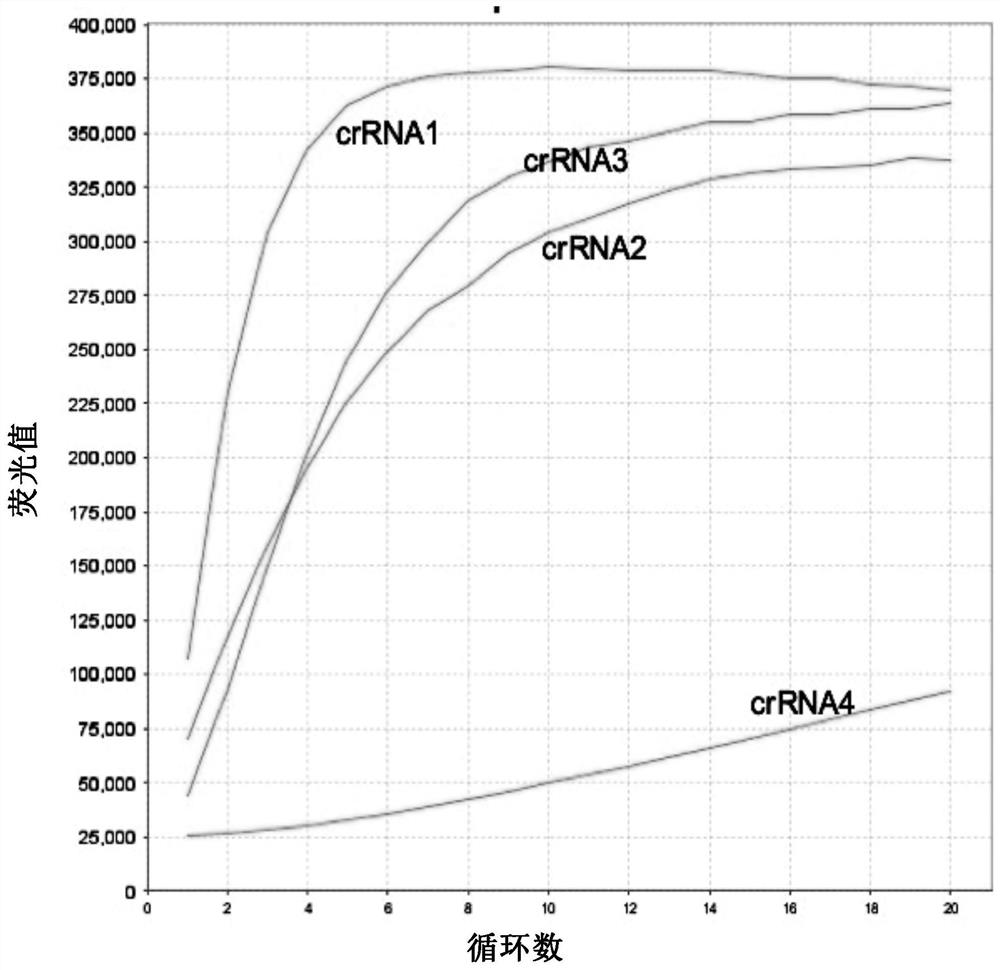
[0198] sequence (5'-3') SEQ ID No: crRNA guide sequence 1: 5'-UGGCUGUAGUUGUGAUCAACUCC-3' 1 crRNA guide sequence 2: 5'-AUCACAACUACAGCCAUAAC-3' 2 crRNA guide sequence 3: 5'-GCGGAGUUGAUCACAACUACAGC-3' 3 crRNA guide sequence 4: 5'-AAAACACAGUCUGUACCGUCUGC-3' 4
[0199] To prepare crRNA probes, T7-crRNA-F was annealed to synthetic oligonucleotides to prepare transcription templates. Specifi...
Embodiment 2
[0217] In this example, the SARS-CoV-2 RNA standard control was serially diluted with HEK293T total RNA, and the qRT-PCR reaction and SENA sensitization reaction were performed respectively. Negative control and positive control were water and synthetic SARS-CoV-2 ORF1ab fragment respectively; the qRT-PCR amplification primers and probes used were the primers recommended by China CDC (see primer pair 2) and matching Taqman fluorescent probes .
[0218] After qRT-PCR reaction, take 2 μL qRT-PCR reaction solution and add it to 18 μL premixed SENA reaction system (LbaCas12a, crRNA1, FAM-N 12 -BHQ1 and reaction buffer), reacted at 37 degrees for 10 minutes.
[0219] The reaction results of qRT-PCR and SENA are shown in Table 1 below. Among them, the detection sensitivity of qRT-PCR can only detect sample No. 5, while SENA can effectively detect sample No. 8, which is at least 8 times more sensitive than qRT-PCR. Among them, UD indicates that qRT-PCR did not amplify the curve, a...
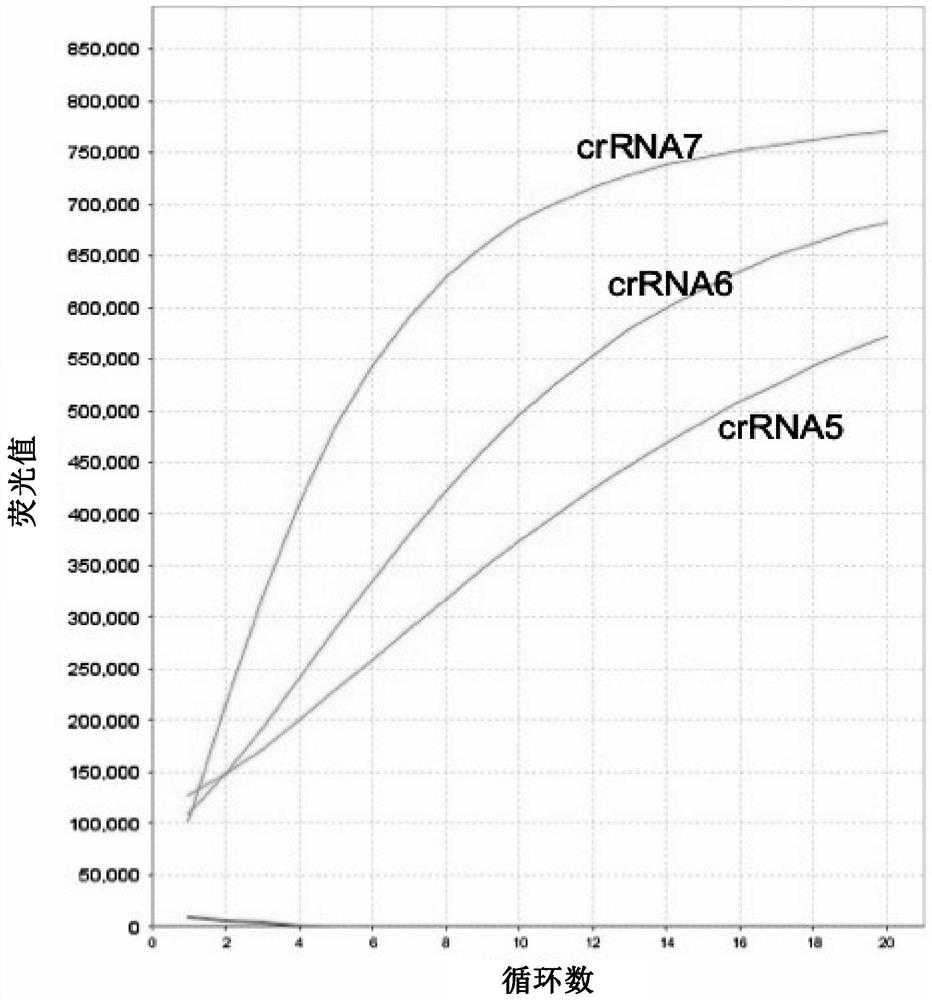
Embodiment 3
[0223] In this example, the synthetic positive control DNA containing the SARS-CoV-2 N gene fragment was serially diluted with human throat swab extract, and qPCR reaction and SENA sensitization reaction were performed respectively. The qPCR amplification primers used are the primers recommended by China CDC (see primer pair 10) and matching Taqman fluorescent probes. After the qPCR reaction, 2 μL of the qPCR reaction solution was added to the pre-mixed 18 μL SENA reaction system (LbaCas12a, crRNA7, FAM-N 12 -BHQ1 and reaction buffer), reacted at 37 degrees for 10 minutes.
[0224] The results of qPCR and SENA sensitization experiments were performed on the positive control samples after serial dilution, respectively, as shown in Table 2 below. When the target DNA molecules in the qPCR reaction system are less than 32, qPCR cannot obtain a stable and effective amplification Ct value. In contrast, the detection limit of SENA can reach 2 target nucleic acid molecules, that is,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com