Receptor reagent for detecting novel coronavirus and application thereof
A coronavirus, a new type of technology, applied in the field of immune detection, can solve the problems of high technical complexity, delayed disease, and long detection window period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Example 1: Preparation of acceptor microspheres
[0085] 1.1 Synthesis of polystyrene latex microspheres
[0086] Prepare a 100mL three-neck flask, add 40mmol styrene, 3mmol methacrylic acid, and 10mL water, stir for 10min, and pass N 2 30min;
[0087] 1) Weigh 0.11g of ammonium persulfate and 0.2g of sodium chloride, dissolve them in 40mL of water to prepare an aqueous solution. This aqueous solution is joined in the reaction system of step 1), continues logical N 2 30min;
[0088] 2) The temperature of the reaction system was raised to 70°C, and the reaction was carried out for 15 hours;
[0089] 3) Cool the emulsion after the reaction to room temperature, and filter it with a suitable filter cloth. The obtained emulsion was washed with deionized water for several times by centrifugation and sedimentation until the conductivity of the supernatant at the beginning of centrifugation was close to that of deionized water, then diluted with water, and preserved in t...
Embodiment 2
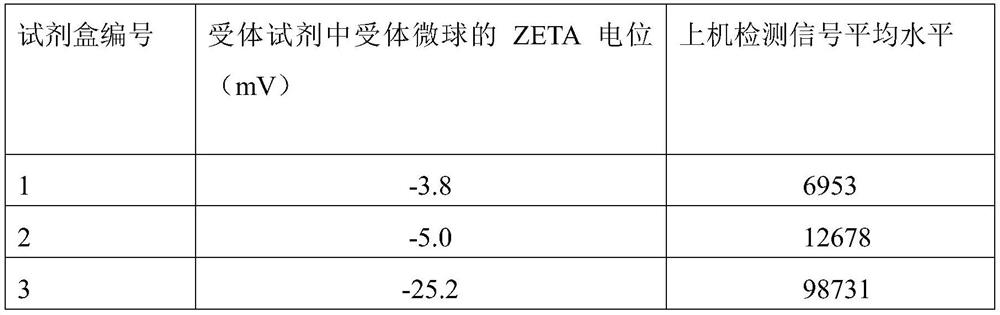
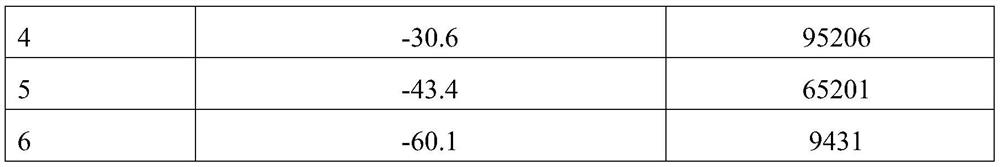
[0105] Embodiment 2: the preparation of kit
[0106] 2.1 Preparation of receptor reagents
[0107] 1) Antibody treatment: dialyze the SARS-CoV-2 spine S1 subunit antibody (or SARS-CoV-2 (2019-nCoV) nucleocapsid protein / N antibody), replace it with coating buffer, and measure the protein concentration;
[0108] 2) Receptor microspheres treatment: the receptor microspheres prepared in Example 1 were replaced by coating buffer through processes such as centrifugation and ultrasound;
[0109] 3) Coupling: The treated receptor microspheres and the treated 2019-nCoV antibody 1 are mixed and reacted. After reduction and blocking, the receptor microspheres-2019-nCoV antibody 1 is obtained, and the preservation solution is used for determination. Contain and store to obtain the receptor reagent.
[0110] 2.2 Preparation of capture reagents
[0111] 1) Antibody treatment: dialyze the 2019-nCoV spike antibody (or 2019-nCoV nucleocapsid protein / N antibody), replace it with labeling bu...
Embodiment 3
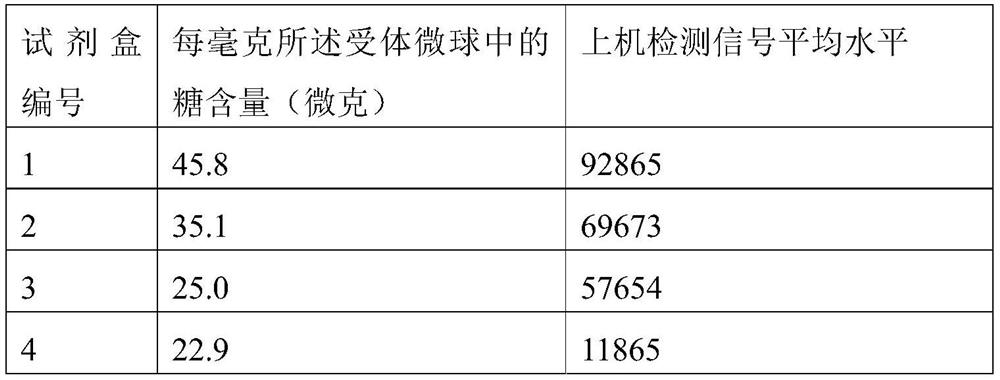
[0115] Embodiment 3: Utilize anthrone method to detect sugar content
[0116] 3.1 Microsphere sample pretreatment:
[0117] The receptor reagents containing 1 mg of receptor microspheres in Example 2 were centrifuged at 20,000 G for 40 min, the supernatant was poured off, and ultrasonically dispersed with purified water. After repeated centrifugation and dispersion three times, each was dilute to 1 mg / mL with purified water as the preparation Check samples.
[0118] 3.2 Preparation of glucose standard solution:
[0119] Prepare 1 mg / mL glucose stock solution with purified water to prepare standard solution curves of 0 mg / mL, 0.025 mg / mL, 0.05 mg / mL, 0.075 mg / mL, 0.10 mg / mL, and 0.15 mg / mL.
[0120] 3.3 Preparation of anthrone solution: Prepare 2 mg / mL with 80% sulfuric acid solution.
[0121] 3.4 Add 0.1mL glucose standard solution of each concentration and the sample to be tested to the centrifuge tube, and add 1mL anthrone test solution to each tube.
[0122] 3.5 Incubat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
| coefficient of variation | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com