Ribosome DNA oligonucleotide mixed probe sleeve based on reference genome of chrysanthemum related species plant and development method
An oligonucleotide probe and reference genome technology, applied in the field of molecular cytogenetics research, can solve the problems of cumbersome probe preparation method and process, signal interference, difficult identification, etc., and achieve outstanding practicability and scientificity, repeatability High-performance, easy-to-detect effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Embodiment 1 Chrysanthemum 5S rDNA and 45S rDNA oligonucleotide probe development
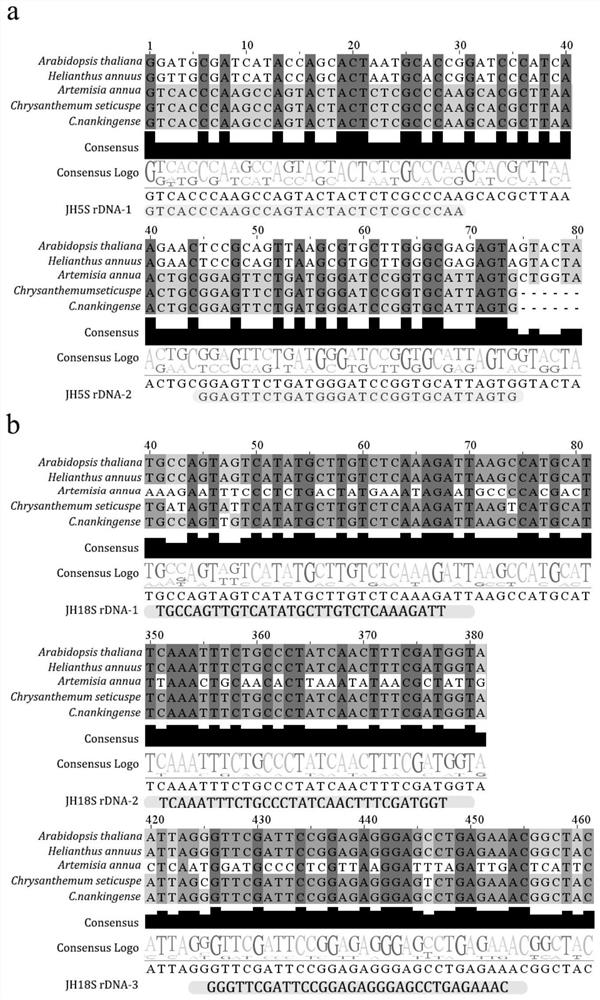
[0053] Using the downloaded Arabidopsis rDNA data, local Blast comparisons were performed on the existing Chrysanthemum Brain, Chamomile, Artemisia annua and Sunflower. The consensus sequence was obtained by comparing 5S, 5.8S, 18S and 28S rDNA sequences respectively, the results are as follows figure 1 shown. The consensus sequence lengths of 5S, 5.8S, 18S and 28S rDNA were 123bp, 164bp, 952bp and 3402bp, respectively. The coding sequence of 5S rDNA showed high similarity in Chrysanthemum chinensis, Chamomile, and Artemisia annua.
[0054] In order to obtain representative 5S rDNA probes, the obtained consensus sequences were used for multiple sequence alignment to design 5S rDNA probes. The comparison results showed that the coding sequence of 5S rDNA was relatively conserved in Chrysanthemum chinensis, Chamomile and Artemisia annua. The synthetic 5SrDNA oligonucleotide probe set ...
Embodiment 2
[0070] Embodiment 2 prepares Chrysanthemum 5S rDNA and 45S rDNA oligonucleotide probe cover hybridization solution
[0071] The oligonucleotide probe set hybridization solution configuration system is as follows:
[0072]
[0073] Further, in the oligonucleotide mixed probe set, the concentration of each probe is JH 5S rDNA-1 (0.55ng / μL); JH 5S rDNA-2 (0.55ng / μL); JH 5.8S rDNA-1 ( 0.55ng / μL); JH 5.8S rDNA-2 (0.55ng / μL); JH 18S rDNA-1 (0.55ng / μL); JH 18S rDNA-2 (0.55ng / μL); JH 18S rDNA-3 (0.55 ng / μL); JH 28S rDNA-1 (0.55ng / μL); JH 28S rDNA-2 (0.55ng / μL); JH 28S rDNA-3 (0.55ng / μL); JH 28S rDNA-4 (0.55ng / μL) μL); JH 28S rDNA-5 (0.55ng / μL). The amount of each probe added was 0.5 μL.
[0074] Mix the above ingredients in a 1.5mL centrifuge tube, shake and centrifuge, place in a heating block at 105°C for denaturation for 13 minutes, take it out, and place it in -20°C ice alcohol for 10 minutes to prepare the oligonucleotide probe set hybridization solution .
Embodiment 3
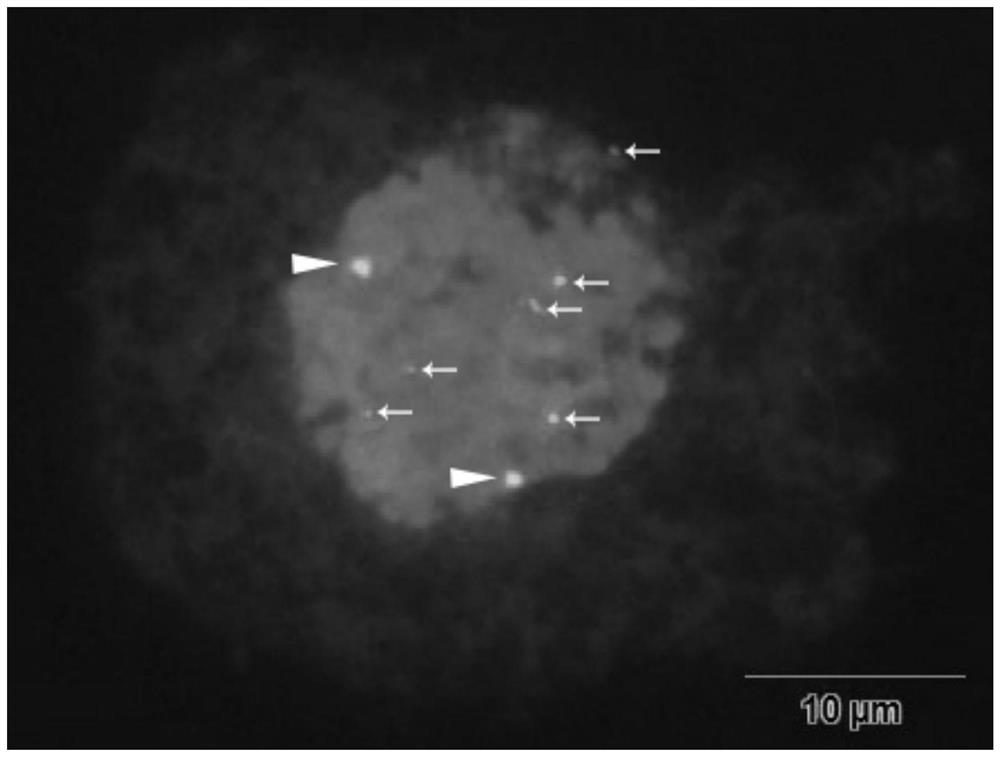
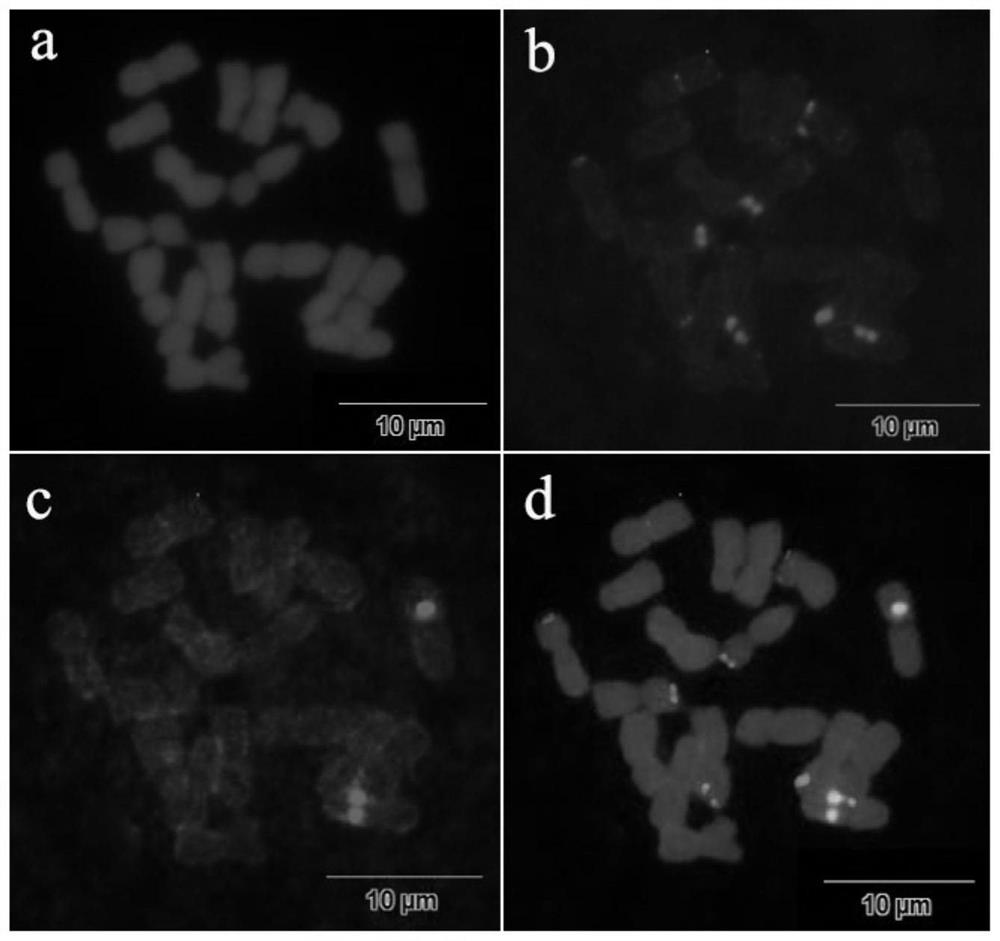
[0075] Example 3 5S rDNA and 45S rDNA oligonucleotide probe set signal detection and high-resolution chromosome fluorescence in situ hybridization of Chrysanthemum plants
[0076] Experimental material: Chrysanthemum brain (C.nankinense) chromosome.
[0077] experiment method:
[0078] (1) Root tip treatment of Chrysanthemum plants: Propagate the experimental materials in MS medium respectively, and take Chrysanthemum plant tissue culture seedlings that have been subcultured 4-5 times in a constant temperature culture room at 25°C to remove residual exogenous substances in the body. Hormone, select the Chrysanthemum tissue culture seedlings after subculture to observe rooting, when 4-5 root systems with a length of 2-3cm appear in the tissue culture seedlings, take out the tissue culture seedlings from the MS medium and wash them thoroughly. Immerse the root system in a petri dish containing 0.2 μmol / L amafonophos-methyl solution for 4 hours, rinse it with clean water, cut of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com